当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Simple and Precise Counting of Viable Bacteria by Resazurin-Amplified Picoarray Detection
Analytical Chemistry ( IF 7.4 ) Pub Date : 2018-07-03 00:00:00 , DOI: 10.1021/acs.analchem.8b02096 Kuangwen Hsieh 1 , Helena C. Zec 2 , Liben Chen 1 , Aniruddha M. Kaushik 1 , Kathleen E. Mach 3 , Joseph C. Liao 3 , Tza-Huei Wang 1, 2
Analytical Chemistry ( IF 7.4 ) Pub Date : 2018-07-03 00:00:00 , DOI: 10.1021/acs.analchem.8b02096 Kuangwen Hsieh 1 , Helena C. Zec 2 , Liben Chen 1 , Aniruddha M. Kaushik 1 , Kathleen E. Mach 3 , Joseph C. Liao 3 , Tza-Huei Wang 1, 2
Affiliation
|
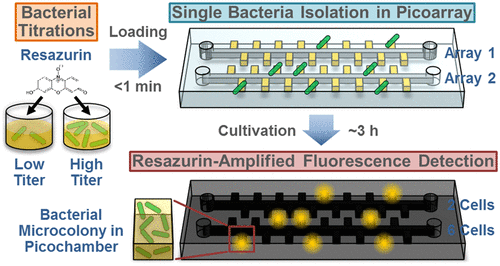
Simple, fast, and precise counting of viable bacteria is fundamental to a variety of microbiological applications such as food quality monitoring and clinical diagnosis. To this end, agar plating, microscopy, and emerging microfluidic devices for single bacteria detection have provided useful means for counting viable bacteria, but they also have their limitations ranging from complexity, time, and inaccuracy. We present herein our new method RAPiD (Resazurin-Amplified Picoarray Detection) for addressing this important problem. In RAPiD, we employ vacuum-assisted sample loading and oil-driven sample digitization to stochastically confine single bacteria in Picoarray, a microfluidic device with picoliter-sized isolation chambers (picochambers), in <30 s with only a few minutes of hands-on time. We add AlamarBlue, a resazurin-based fluorescent dye for bacterial growth, in our assay to accelerate the detection of “microcolonies” proliferated from single bacteria within picochambers. Detecting fluorescence in picochambers as an amplified surrogate for bacterial cells allows us to count hundreds of microcolonies with a single image taken via wide-field fluorescence microscopy. We have also expanded our method to practically test multiple titrations from a single bacterial sample in parallel. Using this expanded “multi-RAPiD” strategy, we can quantify viable cells in E. coli and S. aureus samples with precision in ∼3 h, illustrating RAPiD as a promising new method for counting viable bacteria for microbiological applications.
中文翻译:

通过刃天青素扩增的微阵列检测对活菌进行简单而精确的计数
简单,快速和精确地计算活菌是各种微生物应用(例如食品质量监测和临床诊断)的基础。为此,琼脂平板,显微镜检查和新兴的用于检测单个细菌的微流控设备为计数活菌提供了有用的手段,但它们也有其局限性,包括复杂性,时间和不准确性。我们在本文提出我们的新方法的快速(ř esazurin-甲mplified裨coarray d保护)来解决这个重要问题。在RAPiD中,我们采用真空辅助的样品加载和油驱动的样品数字化技术来随机控制Picoarray(一种具有皮升大小的隔离室(picochambers)的微流控设备)中的单个细菌,在不到30 s的时间内即可完成操作时间。我们在分析中添加了基于刃天青的细菌生长荧光染料AlamarBlue,以加快对皮腔室内单个细菌繁殖的“微菌落”的检测。在微微腔室中检测荧光作为细菌细胞的替代物,使我们能够通过广角荧光显微镜对一张图像进行计数,从而计数数百个微菌落。我们还扩展了我们的方法,以实际从单个细菌样品中并行测试多种滴定度。大肠杆菌和金黄色葡萄球菌样品的精密度约为3小时,说明RAPiD是一种有前途的新方法,可用于计数微生物应用中的活菌。
更新日期:2018-07-03
中文翻译:

通过刃天青素扩增的微阵列检测对活菌进行简单而精确的计数
简单,快速和精确地计算活菌是各种微生物应用(例如食品质量监测和临床诊断)的基础。为此,琼脂平板,显微镜检查和新兴的用于检测单个细菌的微流控设备为计数活菌提供了有用的手段,但它们也有其局限性,包括复杂性,时间和不准确性。我们在本文提出我们的新方法的快速(ř esazurin-甲mplified裨coarray d保护)来解决这个重要问题。在RAPiD中,我们采用真空辅助的样品加载和油驱动的样品数字化技术来随机控制Picoarray(一种具有皮升大小的隔离室(picochambers)的微流控设备)中的单个细菌,在不到30 s的时间内即可完成操作时间。我们在分析中添加了基于刃天青的细菌生长荧光染料AlamarBlue,以加快对皮腔室内单个细菌繁殖的“微菌落”的检测。在微微腔室中检测荧光作为细菌细胞的替代物,使我们能够通过广角荧光显微镜对一张图像进行计数,从而计数数百个微菌落。我们还扩展了我们的方法,以实际从单个细菌样品中并行测试多种滴定度。大肠杆菌和金黄色葡萄球菌样品的精密度约为3小时,说明RAPiD是一种有前途的新方法,可用于计数微生物应用中的活菌。



























 京公网安备 11010802027423号
京公网安备 11010802027423号