当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Simulation-guided engineering of an enzyme-powered three dimensional DNA nanomachine for discriminating single nucleotide variants†
Chemical Science ( IF 8.4 ) Pub Date : 2018-07-02 00:00:00 , DOI: 10.1039/c8sc02761g Yongya Li 1 , Guan A Wang 1 , Sean D Mason 1 , Xiaolong Yang 1 , Zechen Yu 1 , Yanan Tang 1, 2 , Feng Li 1, 2
Chemical Science ( IF 8.4 ) Pub Date : 2018-07-02 00:00:00 , DOI: 10.1039/c8sc02761g Yongya Li 1 , Guan A Wang 1 , Sean D Mason 1 , Xiaolong Yang 1 , Zechen Yu 1 , Yanan Tang 1, 2 , Feng Li 1, 2
Affiliation
|
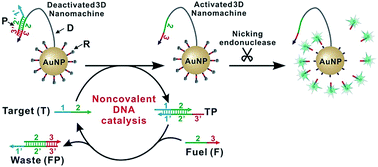
Single nucleotide variants (SNVs) are important both clinically and biologically because of their profound biological consequences. Herein, we engineered a nicking endonuclease-powered three dimensional (3D) DNA nanomachine for discriminating SNVs with high sensitivity and specificity. Particularly, we performed a simulation-guided tuning of sequence designs to achieve the optimal trade-off between device efficiency and specificity. We also introduced an auxiliary probe, a molecular fuel capable of tuning the device in solution via noncovalent catalysis. Collectively, our device produced discrimination factors comparable with commonly used molecular probes but improved the assay sensitivity by ∼100 times. Our results also demonstrate that rationally designed DNA probes through computer simulation can be used to quantitatively improve the design and operation of complexed molecular devices and sensors.
中文翻译:

用于区分单核苷酸变异的酶驱动三维 DNA 纳米机器的模拟引导工程†
单核苷酸变异(SNV)因其深远的生物学后果而在临床和生物学上都很重要。在此,我们设计了一种切口核酸内切酶驱动的三维 (3D) DNA 纳米机器,用于以高灵敏度和特异性区分 SNV。特别是,我们对序列设计进行了模拟引导的调整,以实现设备效率和特异性之间的最佳权衡。我们还引入了一种辅助探针,一种能够通过非共价催化在溶液中调节装置的分子燃料。总的来说,我们的设备产生的辨别因子与常用的分子探针相当,但将测定灵敏度提高了约 100 倍。我们的结果还表明,通过计算机模拟合理设计的 DNA 探针可用于定量改进复杂分子装置和传感器的设计和操作。
更新日期:2018-07-02
中文翻译:

用于区分单核苷酸变异的酶驱动三维 DNA 纳米机器的模拟引导工程†
单核苷酸变异(SNV)因其深远的生物学后果而在临床和生物学上都很重要。在此,我们设计了一种切口核酸内切酶驱动的三维 (3D) DNA 纳米机器,用于以高灵敏度和特异性区分 SNV。特别是,我们对序列设计进行了模拟引导的调整,以实现设备效率和特异性之间的最佳权衡。我们还引入了一种辅助探针,一种能够通过非共价催化在溶液中调节装置的分子燃料。总的来说,我们的设备产生的辨别因子与常用的分子探针相当,但将测定灵敏度提高了约 100 倍。我们的结果还表明,通过计算机模拟合理设计的 DNA 探针可用于定量改进复杂分子装置和传感器的设计和操作。



























 京公网安备 11010802027423号
京公网安备 11010802027423号