当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Elucidating Allosteric Communications in Proteins with Difference Contact Network Analysis
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-06-29 00:00:00 , DOI: 10.1021/acs.jcim.8b00250 Xin-Qiu Yao 1 , Mohamed Momin 1 , Donald Hamelberg 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-06-29 00:00:00 , DOI: 10.1021/acs.jcim.8b00250 Xin-Qiu Yao 1 , Mohamed Momin 1 , Donald Hamelberg 1
Affiliation
|
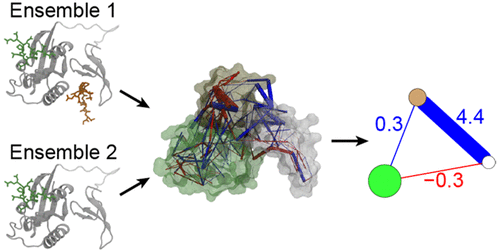
A difference contact network analysis (dCNA) method is developed for delineating allosteric mechanisms in proteins. The new method addresses limitations of conventional network analysis methods and is particularly suitable for allosteric systems undergoing large-amplitude conformational changes during function. Tests show that dCNA works well for proteins of varying sizes and functions. The design of dCNA is general enough to facilitate analyses of diverse dynamic data generated by molecular dynamics, crystallography, or nuclear magnetic resonance.
中文翻译:

通过差异接触网络分析阐明蛋白质的变构通讯
已开发出差异接触网络分析(dCNA)方法来描述蛋白质中的变构机制。该新方法解决了传统网络分析方法的局限性,特别适用于在功能过程中经历大幅度构象变化的变构系统。测试表明,dCNA适用于各种大小和功能的蛋白质。dCNA的设计足够通用,可以方便地分析由分子动力学,晶体学或核磁共振产生的各种动态数据。
更新日期:2018-06-29
中文翻译:

通过差异接触网络分析阐明蛋白质的变构通讯
已开发出差异接触网络分析(dCNA)方法来描述蛋白质中的变构机制。该新方法解决了传统网络分析方法的局限性,特别适用于在功能过程中经历大幅度构象变化的变构系统。测试表明,dCNA适用于各种大小和功能的蛋白质。dCNA的设计足够通用,可以方便地分析由分子动力学,晶体学或核磁共振产生的各种动态数据。



























 京公网安备 11010802027423号
京公网安备 11010802027423号