Biochimie ( IF 3.9 ) Pub Date : 2018-06-27 , DOI: 10.1016/j.biochi.2018.06.017 Sebastien Desbois , Ulrik P. John , Matthew A. Perugini
|
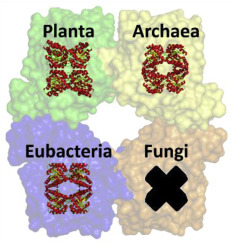
The class I aldolase dihydrodipicolinate synthase (DHDPS) catalyzes the first committed step of the diaminopimelate (DAP) lysine biosynthesis pathway in bacteria, archaea and plants. Despite the existence, in databases, of numerous fungal sequences annotated as DHDPS, its presence in fungi has been the subject of contradictory claims. We report the characterization of DHDPS candidates from fungi. Firstly, the putative DHDPS from Coccidioides immitis (PDB ID: 3QFE) was shown to have negligible enzyme activity. Sequence analysis of 3QFE showed that three out of the seven amino acid residues critical for DHDPS activity are absent; however, exact matches to catalytic residues from two other class I aldolases, 2-keto-3-deoxygluconate aldolase (KDGA), and 4-hydroxy-2-oxoglutarate aldolase (HOGA), were identified. The presence of both KDGA and HOGA activity in 3QFE was confirmed in vitro using enzyme assays, the first report of such dual activity. Subsequent analyses of all publically available fungal sequences revealed that no entry contains all seven residues important for DHDPS function. The candidate with the highest number of identities (6 of 7), KIW77228 from Fonsecaea pedrosoi, was shown to have trace DHDPS activity in vitro, partially restored by substitution of the seventh critical residue, and to be incapable of complementing DHDPS-deficient E. coli cells. Combined with the presence of all seven sequences for the alternative α-aminoadipate (AAA) lysine biosynthesis pathway in C. immitis and F. pedrosoi, we believe that DHDPS and the DAP pathway are absent in fungi, and further, that robust informed methods for annotating genes need to be implemented.
中文翻译:

真菌中不存在二氢二吡啶甲酸合酶
I类醛缩酶二氢二吡啶甲酸合酶(DHDPS)催化细菌,古细菌和植物中二氨基庚二酸(DAP)赖氨酸生物合成途径的第一步。尽管在数据库中存在许多标注为DHDPS的真菌序列,但其在真菌中的存在一直是相互矛盾的主题。我们报告了真菌中DHDPS候选者的特征。首先,从球虫免疫炎中推定的DHDPS(PDB ID:3QFE)具有微不足道的酶活性。3QFE的序列分析表明,对DHDPS活性至关重要的7个氨基酸残基中不存在3个。但是,已确定与其他两个I类醛缩酶,2-酮-3-脱氧葡萄糖酸酯醛缩酶(KDGA)和4-羟基-2-氧戊二酸酯醛缩酶(HOGA)的催化残基完全匹配。使用酶测定法在体外证实了3QFE中同时存在KDGA和HOGA活性,这是此类双重活性的首次报道。随后对所有公共真菌序列的分析表明,没有条目包含对DHDPS功能重要的所有七个残基。具有最高身份标识的候选人(6之7),来自Fonsecaea pedrosoi的KIW77228被证明具有痕量DHDPS活性在体外,通过取代第七个关键残基部分恢复,并且无法补充DHDPS缺陷的大肠杆菌细胞。结合存在于C. immitis和F. pedrosoi中的7种替代α-氨基己二酸(AAA)赖氨酸生物合成途径的全部序列,我们认为DHDPS和DAP途径在真菌中不存在,而且,可靠的方法可用于注释基因需要实现。



























 京公网安备 11010802027423号
京公网安备 11010802027423号