当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Amino Acid Selective 13C Labeling and 13C Scrambling Profile Analysis of Protein α and Side-Chain Carbons in Escherichia coli Utilized for Protein Nuclear Magnetic Resonance
Biochemistry ( IF 2.9 ) Pub Date : 2018-06-20 00:00:00 , DOI: 10.1021/acs.biochem.8b00182 Toshihiko Sugiki 1 , Kyoko Furuita 1 , Toshimichi Fujiwara 1 , Chojiro Kojima 1, 2
Biochemistry ( IF 2.9 ) Pub Date : 2018-06-20 00:00:00 , DOI: 10.1021/acs.biochem.8b00182 Toshihiko Sugiki 1 , Kyoko Furuita 1 , Toshimichi Fujiwara 1 , Chojiro Kojima 1, 2
Affiliation
|
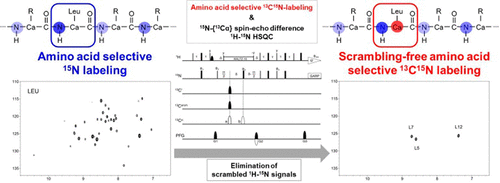
Amino acid selective isotope labeling is an important nuclear magnetic resonance technique, especially for larger proteins, providing strong bases for the unambiguous resonance assignments and information concerning the structure, dynamics, and intermolecular interactions. Amino acid selective 15N labeling suffers from isotope dilution caused by metabolic interconversion of the amino acids, resulting in isotope scrambling within the target protein. Carbonyl 13C atoms experience less isotope scrambling than the main-chain 15N atoms do. However, little is known about the side-chain 13C atoms. Here, the 13C scrambling profiles of the Cα and side-chain carbons were investigated for 15N scrambling-prone amino acids, such as Leu, Ile, Tyr, Phe, Thr, Val, and Ala. The level of isotope scrambling was substantially lower in 13Cα and 13C side-chain labeling than in 15N labeling. We utilized this reduced scrambling-prone character of 13C as a simple and efficient method for amino acid selective 13C labeling using an Escherichia coli cold-shock expression system and high-cell density fermentation. Using this method, the 13C labeling efficiency was >80% for Leu and Ile, ∼60% for Tyr and Phe, ∼50% for Thr, ∼40% for Val, and 30–40% for Ala. 1H–15N heteronuclear single-quantum coherence signals of the 15N scrambling-prone amino acid were also easily filtered using 15N-{13Cα} spin–echo difference experiments. Our method could be applied to the assignment of the 55 kDa protein.
中文翻译:

用于蛋白质核磁共振的大肠杆菌中蛋白质α和侧链碳的氨基酸选择性13 C标记和13 C加扰曲线分析
氨基酸选择性同位素标记是一种重要的核磁共振技术,特别是对于较大的蛋白质,为明确的共振分配以及有关结构,动力学和分子间相互作用的信息提供了坚实的基础。氨基酸选择性15 N标记遭受氨基酸代谢互变引起的同位素稀释,导致目标蛋白内的同位素加扰。羰基13 C原子比主链15 N原子经历更少的同位素加扰。然而,关于侧链13 C原子知之甚少。在这里,研究了15个Cα和侧链碳的13 C加扰曲线Ñ扰频发的氨基酸,如亮氨酸,异亮氨酸,酪氨酸,苯丙氨酸,苏氨酸,缬氨酸,和Ala。同位素的水平加扰中被显着降低13 Cα和13 C侧链标记比在15 Ñ标记。我们利用这种降低的13 C易扰性特征作为使用大肠杆菌冷休克表达系统和高细胞密度发酵进行氨基酸选择性13 C标记的简单有效方法。使用这种方法,Leu和Ile的13 C标记效率> 80%,Tyr和Phe的〜60%,Thr的〜50%,Val的〜40%,Ala的30–40%,1 H– 15所述N个异核单量子相干信号15 Ñ扰频发氨基酸也容易使用过滤15 N- { 13 Cα}自旋回波差实验。我们的方法可以应用于55 kDa蛋白的分配。
更新日期:2018-06-20
中文翻译:

用于蛋白质核磁共振的大肠杆菌中蛋白质α和侧链碳的氨基酸选择性13 C标记和13 C加扰曲线分析
氨基酸选择性同位素标记是一种重要的核磁共振技术,特别是对于较大的蛋白质,为明确的共振分配以及有关结构,动力学和分子间相互作用的信息提供了坚实的基础。氨基酸选择性15 N标记遭受氨基酸代谢互变引起的同位素稀释,导致目标蛋白内的同位素加扰。羰基13 C原子比主链15 N原子经历更少的同位素加扰。然而,关于侧链13 C原子知之甚少。在这里,研究了15个Cα和侧链碳的13 C加扰曲线Ñ扰频发的氨基酸,如亮氨酸,异亮氨酸,酪氨酸,苯丙氨酸,苏氨酸,缬氨酸,和Ala。同位素的水平加扰中被显着降低13 Cα和13 C侧链标记比在15 Ñ标记。我们利用这种降低的13 C易扰性特征作为使用大肠杆菌冷休克表达系统和高细胞密度发酵进行氨基酸选择性13 C标记的简单有效方法。使用这种方法,Leu和Ile的13 C标记效率> 80%,Tyr和Phe的〜60%,Thr的〜50%,Val的〜40%,Ala的30–40%,1 H– 15所述N个异核单量子相干信号15 Ñ扰频发氨基酸也容易使用过滤15 N- { 13 Cα}自旋回波差实验。我们的方法可以应用于55 kDa蛋白的分配。



























 京公网安备 11010802027423号
京公网安备 11010802027423号