当前位置:
X-MOL 学术
›
Chem. Bio. Drug Des.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Insight into structural requirements of antiamoebic flavonoids: 3D‐QSAR and G‐QSAR studies
Chemical Biology & Drug Design ( IF 3 ) Pub Date : 2018-06-26 , DOI: 10.1111/cbdd.13343 Shehnaz Fatima 1 , Payal Gupta 1 , Subhash Mohan Agarwal 1
Chemical Biology & Drug Design ( IF 3 ) Pub Date : 2018-06-26 , DOI: 10.1111/cbdd.13343 Shehnaz Fatima 1 , Payal Gupta 1 , Subhash Mohan Agarwal 1
Affiliation
|
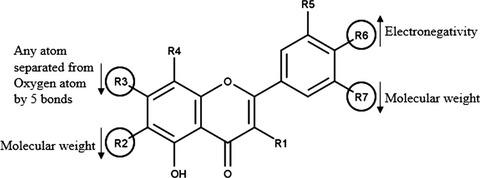
Plant‐based flavonoids have been found to exhibit strong inhibitory capability against Entamoeba histolytica. So, various QSAR models have been developed to identify the critical features that are responsible for the potency of these molecules. 3D‐QSAR analysis using k‐nearest neighbour molecular field analysis via stepwise forward–backward variable selection method showed best results for both internal and external predictive ability of the model (i.e., q2 = 0.64 and pred_r2 = 0.56). Also, a group‐based QSAR (G‐QSAR) model was developed based on partial least squares regression combined with stepwise forward–backward variable selection method. It gave best parametric results (r2 = 0.74, q2 = 0.56 and pred_r2 = 0.54) which implied that the model is highly predictive. 3D‐QSAR established that presence/absence of bulk near rings B and C is important in deciding the inhibitory potential of these molecules. Additionally, G‐QSAR provided site‐specific clue wherein modifications related to molecular weight, electronegativity and separation of an oxygen atom in rings A and C can result in enhanced biological activity. To the best of the author's knowledge, this is the first QSAR study of antiamoebic flavonoids, and therefore, we expect the results to be useful in the design of more potent antiamoebic inhibitors.
中文翻译:

深入了解抗厌氧类黄酮的结构要求:3D-QSAR和G-QSAR研究
已发现基于植物的类黄酮对溶组织变形虫具有很强的抑制能力。因此,已经开发出各种QSAR模型来识别导致这些分子效力的关键特征。通过逐步向前-向后变量选择方法使用k最近邻分子场分析进行的3D-QSAR分析显示了模型内部和外部预测能力的最佳结果(即,q 2 = 0.64和pred_ r 2 = 0.56)。此外,基于偏最小二乘回归结合逐步前向后向变量选择方法,开发了基于组的QSAR(G-QSAR)模型。它给出了最佳的参数结果(r 2 = 0.74,q 2 = 0.56和pred_ r 2 = 0.54),这表明该模型具有高度预测性。3D-QSAR确定B和C环附近是否存在大量物质对于确定这些分子的抑制潜力很重要。此外,G-QSAR提供了特定位置的线索,其中与分子量,电负性和环A和C中氧原子的分离有关的修饰可以增强生物活性。据作者所知,这是首次对抗厌氧类黄酮进行QSAR研究,因此,我们希望该结果可用于设计更有效的抗厌氧抑制剂。
更新日期:2018-06-26
中文翻译:

深入了解抗厌氧类黄酮的结构要求:3D-QSAR和G-QSAR研究
已发现基于植物的类黄酮对溶组织变形虫具有很强的抑制能力。因此,已经开发出各种QSAR模型来识别导致这些分子效力的关键特征。通过逐步向前-向后变量选择方法使用k最近邻分子场分析进行的3D-QSAR分析显示了模型内部和外部预测能力的最佳结果(即,q 2 = 0.64和pred_ r 2 = 0.56)。此外,基于偏最小二乘回归结合逐步前向后向变量选择方法,开发了基于组的QSAR(G-QSAR)模型。它给出了最佳的参数结果(r 2 = 0.74,q 2 = 0.56和pred_ r 2 = 0.54),这表明该模型具有高度预测性。3D-QSAR确定B和C环附近是否存在大量物质对于确定这些分子的抑制潜力很重要。此外,G-QSAR提供了特定位置的线索,其中与分子量,电负性和环A和C中氧原子的分离有关的修饰可以增强生物活性。据作者所知,这是首次对抗厌氧类黄酮进行QSAR研究,因此,我们希望该结果可用于设计更有效的抗厌氧抑制剂。



























 京公网安备 11010802027423号
京公网安备 11010802027423号