npj 2D Materials and Applications ( IF 9.7 ) Pub Date : 2018-05-24 , DOI: 10.1038/s41699-018-0060-8 Amir Barati Farimani , Mohammad Heiranian , Narayana R. Aluru
|
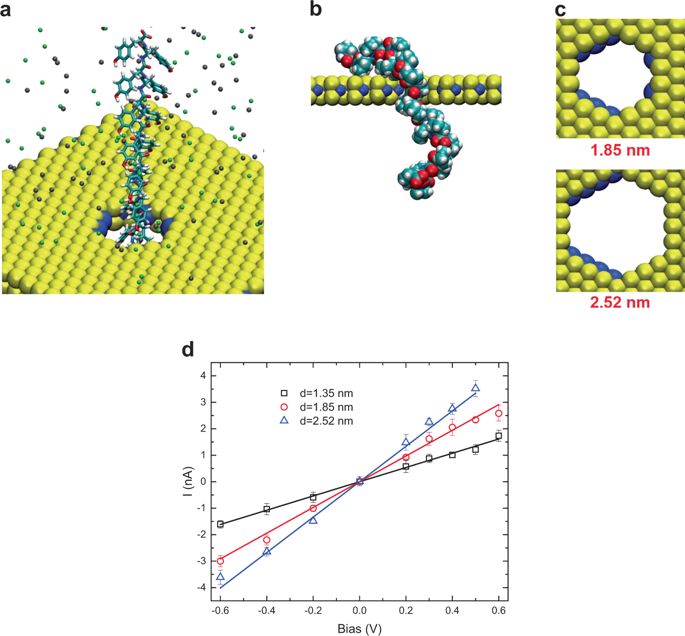
Protein detection plays a key role in determining the single point mutations which can cause a variety of diseases. Nanopore sequencing provides a label-free, single base, fast and long reading platform, which makes it amenable for personalized medicine. A challenge facing nanopore technology is the noise in ionic current. Here, we show that a nanoporous single-layer molybdenum disulfide (MoS2) can detect individual amino acids in a polypeptide chain (16 units) with a high accuracy and distinguishability. Using extensive molecular dynamics simulations (with a total aggregate simulation time of 66 µs) and machine learning techniques, we featurize and cluster the ionic current and residence time of the 20 amino acids and identify the fingerprints of the signals. Using logistic regression, nearest neighbor, and random forest classifiers, the sensor reading is predicted with an accuracy of 72.45, 94.55, and 99.6%, respectively. In addition, using advanced ML classification techniques, we are able to theoretically predict over 2.8 million hypothetical sensor readings’ amino acid types.
中文翻译:

识别具有敏感的纳米多孔MoS 2的氨基酸:面向基于机器学习的预测
蛋白质检测在确定可能导致多种疾病的单点突变中起关键作用。纳米孔测序提供了无标记,单碱基,快速且长读码的平台,使其可用于个性化药物。纳米孔技术面临的挑战是离子电流中的噪声。在这里,我们显示了纳米多孔单层二硫化钼(MoS 2)可以高精度和可区分性检测多肽链中的单个氨基酸(16个单元)。使用广泛的分子动力学模拟(总模拟时间为66 µs)和机器学习技术,我们使20个氨基酸的离子电流和停留时间饱和并聚类,并识别信号的指纹。使用逻辑回归,最近邻和随机森林分类器,传感器读数的预测准确度分别为72.45、94.55和99.6%。此外,使用先进的ML分类技术,我们理论上可以预测280万个假设传感器读数的氨基酸类型。


























 京公网安备 11010802027423号
京公网安备 11010802027423号