当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Engineering a Functional Small RNA Negative Autoregulation Network with Model-Guided Design
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-05-07 00:00:00 , DOI: 10.1021/acssynbio.7b00440 Chelsea Y. Hu 1, 2 , Melissa K. Takahashi 1, 3 , Yan Zhang 1, 4 , Julius B. Lucks 2
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-05-07 00:00:00 , DOI: 10.1021/acssynbio.7b00440 Chelsea Y. Hu 1, 2 , Melissa K. Takahashi 1, 3 , Yan Zhang 1, 4 , Julius B. Lucks 2
Affiliation
|
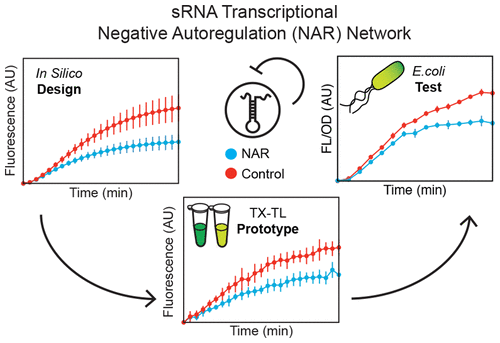
RNA regulators are powerful components of the synthetic biology toolbox. Here, we expand the repertoire of synthetic gene networks built from these regulators by constructing a transcriptional negative autoregulation (NAR) network out of small RNAs (sRNAs). NAR network motifs are core motifs of natural genetic networks, and are known for reducing network response time and steady state signal. Here we use cell-free transcription–translation (TX-TL) reactions and a computational model to design and prototype sRNA NAR constructs. Using parameter sensitivity analysis, we design a simple set of experiments that allow us to accurately predict NAR function in TX-TL. We transfer successful network designs into Escherichia coli and show that our sRNA transcriptional network reduces both network response time and steady-state gene expression. This work broadens our ability to construct increasingly sophisticated RNA genetic networks with predictable function.
中文翻译:

通过模型指导设计工程化的功能性小RNA负自动调节网络
RNA调节剂是合成生物学工具箱的强大组件。在这里,我们通过从小RNA(sRNA)构建转录负自调控(NAR)网络,扩展了由这些调控子构建的合成基因网络的功能。NAR网络主题是自然遗传网络的核心主题,众所周知,它可以减少网络响应时间和稳态信号。在这里,我们使用无细胞转录-翻译(TX-TL)反应和计算模型来设计和原型化sRNA NAR构建体。使用参数敏感性分析,我们设计了一组简单的实验,使我们能够准确预测TX-TL中的NAR功能。我们将成功的网络设计转移到大肠杆菌中并表明我们的sRNA转录网络减少了网络响应时间和稳态基因表达。这项工作拓宽了我们构建具有可预测功能的日益复杂的RNA遗传网络的能力。
更新日期:2018-05-07
中文翻译:

通过模型指导设计工程化的功能性小RNA负自动调节网络
RNA调节剂是合成生物学工具箱的强大组件。在这里,我们通过从小RNA(sRNA)构建转录负自调控(NAR)网络,扩展了由这些调控子构建的合成基因网络的功能。NAR网络主题是自然遗传网络的核心主题,众所周知,它可以减少网络响应时间和稳态信号。在这里,我们使用无细胞转录-翻译(TX-TL)反应和计算模型来设计和原型化sRNA NAR构建体。使用参数敏感性分析,我们设计了一组简单的实验,使我们能够准确预测TX-TL中的NAR功能。我们将成功的网络设计转移到大肠杆菌中并表明我们的sRNA转录网络减少了网络响应时间和稳态基因表达。这项工作拓宽了我们构建具有可预测功能的日益复杂的RNA遗传网络的能力。


























 京公网安备 11010802027423号
京公网安备 11010802027423号