当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Chemical-Mediated Digestion: An Alternative Realm for Middle-down Proteomics?
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2018-05-18 , DOI: 10.1021/acs.jproteome.7b00834 Kristina Srzentić 1 , Konstantin O. Zhurov 1 , Anna A. Lobas 2 , Gennady Nikitin 1 , Luca Fornelli 1 , Mikhail V. Gorshkov 2, 3 , Yury O. Tsybin 4
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2018-05-18 , DOI: 10.1021/acs.jproteome.7b00834 Kristina Srzentić 1 , Konstantin O. Zhurov 1 , Anna A. Lobas 2 , Gennady Nikitin 1 , Luca Fornelli 1 , Mikhail V. Gorshkov 2, 3 , Yury O. Tsybin 4
Affiliation
|
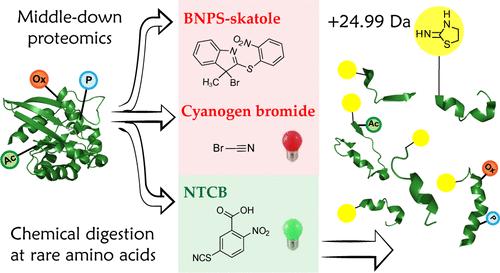
Protein digestion in mass spectrometry (MS)-based bottom-up proteomics targets mainly lysine and arginine residues, yielding primarily 0.6–3 kDa peptides for the proteomes of organisms of all major kingdoms. Recent advances in MS technology enable analysis of complex mixtures of increasingly longer (>3 kDa) peptides in a high-throughput manner supporting the development of a middle-down proteomics (MDP) approach. Generating longer peptides is a paramount step in launching an MDP pipeline, but the quest for the selection of a cleaving agent that would provide the desired 3–15 kDa peptides remains open. Recent bioinformatics studies have shown that cleavage at the rarely occurring amino acid residues such as methionine (Met), tryptophan (Trp), or cysteine (Cys) would be suitable for MDP approach. Interestingly, chemical-mediated proteolytic cleavages uniquely allow targeting these rare amino acids, for which no specific proteolytic enzymes are known. Herein, as potential candidates for MDP-grade proteolysis, we have investigated the performance of chemical agents previously reported to target primarily Met, Trp, and Cys residues: CNBr, BNPS-Skatole (3-bromo-3-methyl-2-(2-nitrophenyl)sulfanylindole), and NTCB (2-nitro-5-thiobenzoic acid), respectively. Figures of merit such as digestion reproducibility, peptide size distribution, and occurrence of side reactions are discussed. The NTCB-based MDP workflow has demonstrated particularly attractive performance, and NTCB is put forward here as a potential cleaving agent for further MDP development.
中文翻译:

化学介导的消化:中下蛋白质组学的替代领域?
基于质谱(MS)的自下而上的蛋白质组学中的蛋白质消化主要针对赖氨酸和精氨酸残基,主要产生用于所有主要王国生物体蛋白质组的0.6–3 kDa肽。MS技术的最新进展使得能够以高通量的方式分析越来越长的(> 3 kDa)肽的复杂混合物,从而支持了中下蛋白质组学(MDP)方法的发展。生成更长的肽段是启动MDP流程的最重要步骤,但是寻求提供所需3-15 kDa肽段的裂解剂的选择仍未解决。最近的生物信息学研究表明,在蛋氨酸(Met),色氨酸(Trp)或半胱氨酸(Cys)等罕见氨基酸残基处进行裂解将适用于MDP方法。有趣的是,化学介导的蛋白水解切割独特地允许靶向这些稀有氨基酸,而对于这些稀有氨基酸而言,尚无特异性蛋白水解酶。在这里,作为MDP级蛋白水解的潜在候选者,我们研究了先前报道的主要针对Met,Trp和Cys残基的化学试剂的性能:CNBr,BNPS-Skatole(3-bromo-3-methyl-2-(2 -硝基苯基)磺酰亚氨基)和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。作为MDP级蛋白水解的潜在候选者,我们研究了先前报道的主要针对Met,Trp和Cys残基的化学试剂的性能:CNBr,BNPS-Skatole(3-bromo-3-methyl-2-(2-nitrophenyl )磺酰亚胺)和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。作为MDP级蛋白水解的潜在候选者,我们研究了先前报道的主要针对Met,Trp和Cys残基的化学试剂的性能:CNBr,BNPS-Skatole(3-bromo-3-methyl-2-(2-nitrophenyl )磺酰亚胺)和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。
更新日期:2018-05-18
中文翻译:

化学介导的消化:中下蛋白质组学的替代领域?
基于质谱(MS)的自下而上的蛋白质组学中的蛋白质消化主要针对赖氨酸和精氨酸残基,主要产生用于所有主要王国生物体蛋白质组的0.6–3 kDa肽。MS技术的最新进展使得能够以高通量的方式分析越来越长的(> 3 kDa)肽的复杂混合物,从而支持了中下蛋白质组学(MDP)方法的发展。生成更长的肽段是启动MDP流程的最重要步骤,但是寻求提供所需3-15 kDa肽段的裂解剂的选择仍未解决。最近的生物信息学研究表明,在蛋氨酸(Met),色氨酸(Trp)或半胱氨酸(Cys)等罕见氨基酸残基处进行裂解将适用于MDP方法。有趣的是,化学介导的蛋白水解切割独特地允许靶向这些稀有氨基酸,而对于这些稀有氨基酸而言,尚无特异性蛋白水解酶。在这里,作为MDP级蛋白水解的潜在候选者,我们研究了先前报道的主要针对Met,Trp和Cys残基的化学试剂的性能:CNBr,BNPS-Skatole(3-bromo-3-methyl-2-(2 -硝基苯基)磺酰亚氨基)和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。作为MDP级蛋白水解的潜在候选者,我们研究了先前报道的主要针对Met,Trp和Cys残基的化学试剂的性能:CNBr,BNPS-Skatole(3-bromo-3-methyl-2-(2-nitrophenyl )磺酰亚胺)和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。作为MDP级蛋白水解的潜在候选者,我们研究了先前报道的主要针对Met,Trp和Cys残基的化学试剂的性能:CNBr,BNPS-Skatole(3-bromo-3-methyl-2-(2-nitrophenyl )磺酰亚胺)和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。和NTCB(2-硝基-5-硫代苯甲酸)。讨论了诸如消化重现性,肽大小分布和副反应发生等优点。基于NTCB的MDP工作流程表现出了特别吸引人的性能,NTCB在这里被提出作为进一步MDP开发的潜在分裂剂。


























 京公网安备 11010802027423号
京公网安备 11010802027423号