当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Predictive Model of Linear Antimicrobial Peptides Active against Gram-Negative Bacteria
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-05-02 00:00:00 , DOI: 10.1021/acs.jcim.8b00118 Boris Vishnepolsky 1 , Andrei Gabrielian 2 , Alex Rosenthal 2 , Darrell E. Hurt 2 , Michael Tartakovsky 2 , Grigol Managadze 1 , Maya Grigolava 1 , George I. Makhatadze 3 , Malak Pirtskhalava 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-05-02 00:00:00 , DOI: 10.1021/acs.jcim.8b00118 Boris Vishnepolsky 1 , Andrei Gabrielian 2 , Alex Rosenthal 2 , Darrell E. Hurt 2 , Michael Tartakovsky 2 , Grigol Managadze 1 , Maya Grigolava 1 , George I. Makhatadze 3 , Malak Pirtskhalava 1
Affiliation
|
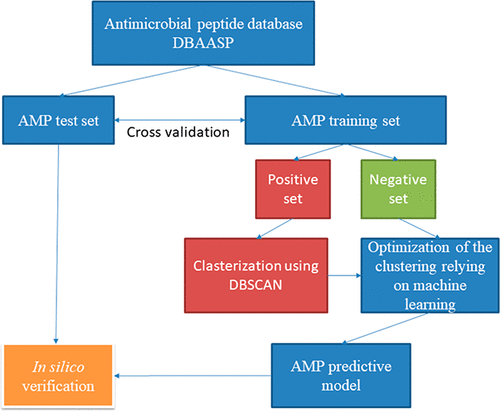
Antimicrobial peptides (AMPs) have been identified as a potential new class of anti-infectives for drug development. There are a lot of computational methods that try to predict AMPs. Most of them can only predict if a peptide will show any antimicrobial potency, but to the best of our knowledge, there are no tools which can predict antimicrobial potency against particular strains. Here we present a predictive model of linear AMPs being active against particular Gram-negative strains relying on a semi-supervised machine-learning approach with a density-based clustering algorithm. The algorithm can well distinguish peptides active against particular strains from others which may also be active but not against the considered strain. The available AMP prediction tools cannot carry out this task. The prediction tool based on the algorithm suggested herein is available on https://dbaasp.org
中文翻译:

对革兰氏阴性细菌有效的线性抗菌肽的预测模型
抗菌肽(AMPs)已被确定为药物开发的潜在新型抗感染剂。有许多计算方法可以尝试预测AMP。他们中的大多数只能预测某肽是否会显示出任何抗菌效力,但据我们所知,尚无工具可以预测针对特定菌株的抗菌效力。在这里,我们提出了一种线性AMP的预测模型,该模型依赖于基于密度的聚类算法的半监督机器学习方法来对抗特定的革兰氏阴性菌株。该算法可以很好地区分对特定菌株具有活性的肽与对其他菌株也可能具有活性但对所考虑的菌株不具有活性的肽。可用的AMP预测工具无法执行此任务。
更新日期:2018-05-02
中文翻译:

对革兰氏阴性细菌有效的线性抗菌肽的预测模型
抗菌肽(AMPs)已被确定为药物开发的潜在新型抗感染剂。有许多计算方法可以尝试预测AMP。他们中的大多数只能预测某肽是否会显示出任何抗菌效力,但据我们所知,尚无工具可以预测针对特定菌株的抗菌效力。在这里,我们提出了一种线性AMP的预测模型,该模型依赖于基于密度的聚类算法的半监督机器学习方法来对抗特定的革兰氏阴性菌株。该算法可以很好地区分对特定菌株具有活性的肽与对其他菌株也可能具有活性但对所考虑的菌株不具有活性的肽。可用的AMP预测工具无法执行此任务。



























 京公网安备 11010802027423号
京公网安备 11010802027423号