当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Peptidic Macrocycles - Conformational Sampling and Thermodynamic Characterization.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-04-20 , DOI: 10.1021/acs.jcim.8b00097 Anna S Kamenik 1 , Uta Lessel 2 , Julian E Fuchs 3 , Thomas Fox 2 , Klaus R Liedl 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-04-20 , DOI: 10.1021/acs.jcim.8b00097 Anna S Kamenik 1 , Uta Lessel 2 , Julian E Fuchs 3 , Thomas Fox 2 , Klaus R Liedl 1
Affiliation
|
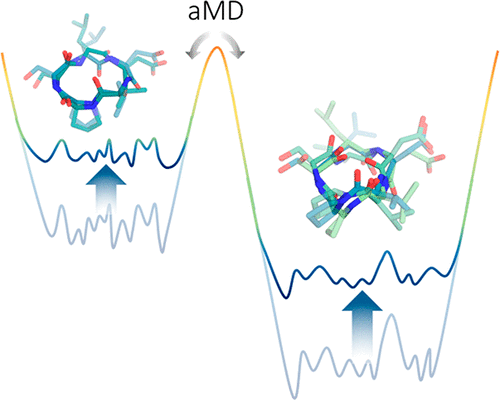
Macrocycles are of considerable interest as highly specific drug candidates, yet they challenge standard conformer generators with their large number of rotatable bonds and conformational restrictions. Here, we present a molecular dynamics-based routine that bypasses current limitations in conformational sampling and extensively profiles the free energy landscape of peptidic macrocycles in solution. We perform accelerated molecular dynamics simulations to capture a diverse conformational ensemble. By applying an energetic cutoff, followed by geometric clustering, we demonstrate the striking robustness and efficiency of the approach in identifying highly populated conformational states of cyclic peptides. The resulting structural and thermodynamic information is benchmarked against interproton distances from NMR experiments and conformational states identified by X-ray crystallography. Using three different model systems of varying size and flexibility, we show that the method reliably reproduces experimentally determined structural ensembles and is capable of identifying key conformational states that include the bioactive conformation. Thus, the described approach is a robust method to generate conformations of peptidic macrocycles and holds promise for structure-based drug design.
中文翻译:

肽大环化合物-构象采样和热力学表征。
大环化合物作为高度特异的候选药物备受关注,但是它们以其大量的可旋转键和构象限制而挑战标准构象异构体产生者。在这里,我们介绍了一个基于分子动力学的例程,该例程绕过了构象采样中的当前限制,并广泛地介绍了肽大环化合物在溶液中的自由能态。我们执行加速的分子动力学模拟,以捕获各种构象集合。通过应用高能截止值,然后进行几何聚类,我们证明了该方法在识别高密度环肽构象状态中的强大鲁棒性和效率。所得的结构和热力学信息以NMR实验中的质子间距和X射线晶体学鉴定的构象状态为基准。使用大小和灵活性不同的三个不同的模型系统,我们表明该方法可靠地重现了实验确定的结构体,并且能够识别包括生物活性构象在内的关键构象状态。因此,所描述的方法是产生肽类大环化合物构象的可靠方法,并有望用于基于结构的药物设计。我们表明该方法可靠地重现了实验确定的结构体,并且能够识别包括生物活性构象在内的关键构象状态。因此,所描述的方法是产生肽类大环化合物构象的可靠方法,并有望用于基于结构的药物设计。我们表明该方法可靠地重现了实验确定的结构体,并且能够识别包括生物活性构象在内的关键构象状态。因此,所描述的方法是产生肽类大环化合物构象的可靠方法,并有望用于基于结构的药物设计。
更新日期:2018-04-13
中文翻译:

肽大环化合物-构象采样和热力学表征。
大环化合物作为高度特异的候选药物备受关注,但是它们以其大量的可旋转键和构象限制而挑战标准构象异构体产生者。在这里,我们介绍了一个基于分子动力学的例程,该例程绕过了构象采样中的当前限制,并广泛地介绍了肽大环化合物在溶液中的自由能态。我们执行加速的分子动力学模拟,以捕获各种构象集合。通过应用高能截止值,然后进行几何聚类,我们证明了该方法在识别高密度环肽构象状态中的强大鲁棒性和效率。所得的结构和热力学信息以NMR实验中的质子间距和X射线晶体学鉴定的构象状态为基准。使用大小和灵活性不同的三个不同的模型系统,我们表明该方法可靠地重现了实验确定的结构体,并且能够识别包括生物活性构象在内的关键构象状态。因此,所描述的方法是产生肽类大环化合物构象的可靠方法,并有望用于基于结构的药物设计。我们表明该方法可靠地重现了实验确定的结构体,并且能够识别包括生物活性构象在内的关键构象状态。因此,所描述的方法是产生肽类大环化合物构象的可靠方法,并有望用于基于结构的药物设计。我们表明该方法可靠地重现了实验确定的结构体,并且能够识别包括生物活性构象在内的关键构象状态。因此,所描述的方法是产生肽类大环化合物构象的可靠方法,并有望用于基于结构的药物设计。



























 京公网安备 11010802027423号
京公网安备 11010802027423号