当前位置:
X-MOL 学术
›
Cell Chem. Bio.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Structural Insights into a Flavin-Dependent [4 + 2] Cyclase that Catalyzes trans-Decalin Formation in Pyrroindomycin Biosynthesis
Cell Chemical Biology ( IF 8.6 ) Pub Date : 2018-04-12 , DOI: 10.1016/j.chembiol.2018.03.007 Qingfei Zheng , Yukang Gong , Yujiao Guo , Zhixiong Zhao , Zhuhua Wu , Zixuan Zhou , Dandan Chen , Lifeng Pan , Wen Liu
Cell Chemical Biology ( IF 8.6 ) Pub Date : 2018-04-12 , DOI: 10.1016/j.chembiol.2018.03.007 Qingfei Zheng , Yukang Gong , Yujiao Guo , Zhixiong Zhao , Zhuhua Wu , Zixuan Zhou , Dandan Chen , Lifeng Pan , Wen Liu
|
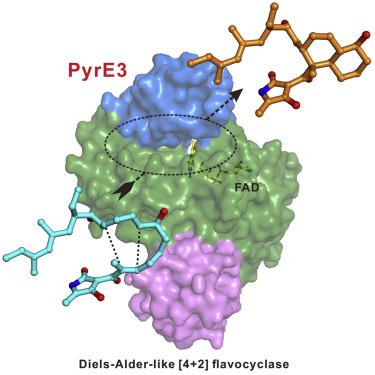
Here, we provide structural insights into PyrE3, a flavin-dependent [4 + 2] cyclase that catalyzestrans-decalin formation in the biosynthesis of pyrroindomycins. PyrE3 shares an architecture/domain organization head-to-tail similarity with the members of the family ofpara-hydroxybenzoate hydroxylase (pHBH)-fold monooxygenases, and possesses a flavin adenine dinucleotide (FAD)-binding domain, a middle domain, and a C-terminal thioredoxin-like domain. The FAD-binding domain forms a central hub of the protein structure, and binds with FAD in a “closed” conformation of pHBH-fold family monooxygenases known for their highly dynamic catalytic processes. FAD plays an essential structural role in PyrE3, where it is amenable to redox change; however, redox change has little effect on [4 + 2] cyclization activity. PyrE3 appears to selectively accommodate a tetramate-containing, linear polyene intermediate in a highly positively charged pocket, which is located at the interface between the FAD-binding domain and the middle domain, and can acceleratetrans-decalin formation likely through anendo-selective [4 + 2] transition state.
中文翻译:

对黄素依赖性[4 + 2]环化酶的结构见解,该环化酶催化吡咯并霉素生物合成中的反式十氢化萘形成
在这里,我们提供对PyrE3的结构性见解,PyrE3是黄素依赖性[4 + 2]环化酶,可在吡咯并霉素的生物合成中催化反式十氢化萘的形成。PyrE3与对羟基苯甲酸酯羟化酶(pHBH)折叠单加氧酶家族成员的构架/结构域结构从头到尾具有相似性,并具有黄素腺嘌呤二核苷酸(FAD)结合结构域,中间结构域和C -末端的硫氧还蛋白样结构域。FAD结合结构域形成蛋白质结构的中心枢纽,并以众所周知的具有高动态催化过程的pHBH折叠家族单加氧酶的“封闭”构型与FAD结合。FAD在PyrE3中起着至关重要的结构作用,在PyrE3中它可以氧化还原变化。但是,氧化还原变化对[4 + 2]环化活性影响很小。
更新日期:2018-06-22
中文翻译:

对黄素依赖性[4 + 2]环化酶的结构见解,该环化酶催化吡咯并霉素生物合成中的反式十氢化萘形成
在这里,我们提供对PyrE3的结构性见解,PyrE3是黄素依赖性[4 + 2]环化酶,可在吡咯并霉素的生物合成中催化反式十氢化萘的形成。PyrE3与对羟基苯甲酸酯羟化酶(pHBH)折叠单加氧酶家族成员的构架/结构域结构从头到尾具有相似性,并具有黄素腺嘌呤二核苷酸(FAD)结合结构域,中间结构域和C -末端的硫氧还蛋白样结构域。FAD结合结构域形成蛋白质结构的中心枢纽,并以众所周知的具有高动态催化过程的pHBH折叠家族单加氧酶的“封闭”构型与FAD结合。FAD在PyrE3中起着至关重要的结构作用,在PyrE3中它可以氧化还原变化。但是,氧化还原变化对[4 + 2]环化活性影响很小。



























 京公网安备 11010802027423号
京公网安备 11010802027423号