PLOS ONE ( IF 3.7 ) Pub Date : 2018-03-16 , DOI: 10.1371/journal.pone.0194630 Davina Gale , Andrew R. J. Lawson , Karen Howarth , Mikidache Madi , Bradley Durham , Sarah Smalley , John Calaway , Shannon Blais , Greg Jones , James Clark , Peter Dimitrov , Michelle Pugh , Samuel Woodhouse , Michael Epstein , Ana Fernandez-Gonzalez , Alexandra S. Whale , Jim F. Huggett , Carole A. Foy , Gerwyn M. Jones , Hadas Raveh-Amit , Karin Schmitt , Alison Devonshire , Emma Green , Tim Forshew , Vincent Plagnol , Nitzan Rosenfeld
|
Introduction
Detection and monitoring of circulating tumor DNA (ctDNA) is rapidly becoming a diagnostic, prognostic and predictive tool in cancer patient care. A growing number of gene targets have been identified as diagnostic or actionable, requiring the development of reliable technology that provides analysis of multiple genes in parallel. We have developed the InVision™ liquid biopsy platform which utilizes enhanced TAm-Seq™ (eTAm-Seq™) technology, an amplicon-based next generation sequencing method for the identification of clinically-relevant somatic alterations at low frequency in ctDNA across a panel of 35 cancer-related genes.
Materials and methods
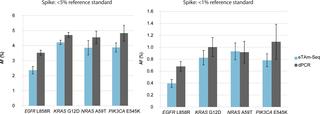
We present analytical validation of the eTAm-Seq technology across two laboratories to determine the reproducibility of mutation identification. We assess the quantitative performance of eTAm-Seq technology for analysis of single nucleotide variants in clinically-relevant genes as compared to digital PCR (dPCR), using both established DNA standards and novel full-process control material.
Results
The assay detected mutant alleles down to 0.02% AF, with high per-base specificity of 99.9997%. Across two laboratories, analysis of samples with optimal amount of DNA detected 94% mutations at 0.25%-0.33% allele fraction (AF), with 90% of mutations detected for samples with lower amounts of input DNA.
Conclusions
These studies demonstrate that eTAm-Seq technology is a robust and reproducible technology for the identification and quantification of somatic mutations in circulating tumor DNA, and support its use in clinical applications for precision medicine.
中文翻译:

开发高度灵敏的液体活检平台,以检测无细胞DNA中低等位基因分数的临床相关癌症突变
介绍
循环肿瘤DNA(ctDNA)的检测和监视正迅速成为癌症患者护理中的诊断,预后和预测工具。越来越多的基因靶标已被确定为可诊断或可操作的,因此需要开发可靠的技术来并行分析多个基因。我们已经开发了InVision™液体活检平台,该平台利用增强的TAm-Seq™(eTAm-Seq™)技术,这是一种基于扩增子的下一代测序方法,可在整个小组的ctDNA中以低频识别临床相关的体细胞改变。 35个与癌症相关的基因。
材料和方法
我们介绍了跨两个实验室对eTAm-Seq技术的分析验证,以确定突变鉴定的可重复性。我们使用既有的DNA标准和新型全过程对照材料,与数字PCR(dPCR)相比,评估了eTAm-Seq技术在临床相关基因中单核苷酸变异体分析的定量性能。
结果
该测定法检测到AF低至0.02%的突变等位基因,具有高达99.9997%的高碱基特异性。在两个实验室中,分析具有最佳DNA量的样品时,在0.25%-0.33%等位基因分数(AF)下检测到94%的突变,而对于输入DNA量较少的样品,则检测到90%的突变。
结论
这些研究表明,eTAm-Seq技术是一种可靠且可重现的技术,用于鉴定和定量循环肿瘤DNA中的体细胞突变,并支持其在精密医学的临床应用中的使用。


























 京公网安备 11010802027423号
京公网安备 11010802027423号