当前位置:
X-MOL 学术
›
ACS Cent. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Defining the Structure of a Protein-Spherical Nucleic Acid Conjugate and Its Counterionic Cloud.
ACS Central Science ( IF 18.2 ) Pub Date : 2018-03-13 , DOI: 10.1021/acscentsci.7b00577 Kurinji Krishnamoorthy 1 , Kyle Hoffmann 2 , Sumit Kewalramani 2 , Jeffrey D Brodin 3 , Liane M Moreau 2 , Chad A Mirkin 2, 3 , Monica Olvera de la Cruz 1, 2, 3, 4 , Michael J Bedzyk 1, 2, 4
ACS Central Science ( IF 18.2 ) Pub Date : 2018-03-13 , DOI: 10.1021/acscentsci.7b00577 Kurinji Krishnamoorthy 1 , Kyle Hoffmann 2 , Sumit Kewalramani 2 , Jeffrey D Brodin 3 , Liane M Moreau 2 , Chad A Mirkin 2, 3 , Monica Olvera de la Cruz 1, 2, 3, 4 , Michael J Bedzyk 1, 2, 4
Affiliation
|
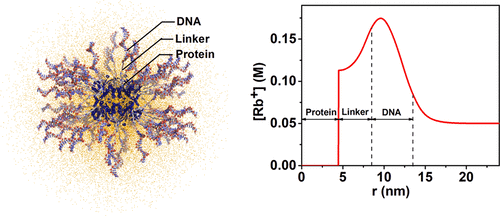
Protein-spherical nucleic acid conjugates (Pro-SNAs) are an emerging class of bioconjugates that have properties defined by their protein cores and dense shell of oligonucleotides. They have been used as building blocks in DNA-driven crystal engineering strategies and show promise as agents that can cross cell membranes and affect both protein and DNA-mediated processes inside cells. However, ionic environments surrounding proteins can influence their activity and conformational stability, and functionalizing proteins with DNA substantively changes the surrounding ionic environment in a nonuniform manner. Techniques typically used to determine protein structure fail to capture such irregular ionic distributions. Here, we determine the counterion radial distribution profile surrounding Pro-SNAs dispersed in RbCl with 1 nm resolution through in situ anomalous small-angle X-ray scattering (ASAXS) and classical density functional theory (DFT). SAXS analysis also reveals the radial extension of the DNA and the linker used to covalently attach the DNA to the protein surface. At the experimental salt concentration of 50 mM RbCl, Rb+ cations compensate ∼90% of the negative charge due to the DNA and linker. Above 75 mM, DFT calculations predict overcompensation of the DNA charge by Rb+. This study suggests a method for exploring Pro-SNA structure and function in different environments through predictions of ionic cloud densities as a function of salt concentration, DNA grafting density, and length. Overall, our study demonstrates that solution X-ray scattering combined with DFT can discern counterionic distribution and submolecular features of highly charged, complex nanoparticle constructs such as Pro-SNAs and related nucleic acid conjugate materials.
中文翻译:

定义蛋白质-球形核酸缀合物的结构及其抗衡离子云。
蛋白质球形核酸偶联物(Pro-SNA)是一类新兴的生物偶联物,其性质由其蛋白质核心和寡核苷酸的致密外壳决定。它们已被用作DNA驱动的晶体工程策略的基本组成部分,并有望作为能穿过细胞膜并影响细胞内蛋白质和DNA介导的过程的试剂。但是,蛋白质周围的离子环境会影响其活性和构象稳定性,并且使用DNA对蛋白质进行功能化会以非均匀的方式实质性地改变周围的离子环境。通常用于确定蛋白质结构的技术无法捕获这种不规则的离子分布。这里,我们通过原位反常小角度X射线散射(ASAXS)和经典密度泛函理论(DFT),确定了以1 nm分辨率分散在RbCl中的Pro-SNA周围的反离子径向分布曲线。SAXS分析还揭示了DNA的径向延伸以及用于将DNA共价附于蛋白质表面的接头。在50 mM RbCl的实验盐浓度下,Rb +阳离子由于DNA和接头而补偿了约90%的负电荷。高于75 mM时,DFT计算可预测Rb +对DNA电荷的过度补偿。这项研究提出了一种通过预测离子云密度与盐浓度,DNA嫁接密度和长度的函数关系来探索Pro-SNA在不同环境中的结构和功能的方法。全面的,
更新日期:2018-03-13
中文翻译:

定义蛋白质-球形核酸缀合物的结构及其抗衡离子云。
蛋白质球形核酸偶联物(Pro-SNA)是一类新兴的生物偶联物,其性质由其蛋白质核心和寡核苷酸的致密外壳决定。它们已被用作DNA驱动的晶体工程策略的基本组成部分,并有望作为能穿过细胞膜并影响细胞内蛋白质和DNA介导的过程的试剂。但是,蛋白质周围的离子环境会影响其活性和构象稳定性,并且使用DNA对蛋白质进行功能化会以非均匀的方式实质性地改变周围的离子环境。通常用于确定蛋白质结构的技术无法捕获这种不规则的离子分布。这里,我们通过原位反常小角度X射线散射(ASAXS)和经典密度泛函理论(DFT),确定了以1 nm分辨率分散在RbCl中的Pro-SNA周围的反离子径向分布曲线。SAXS分析还揭示了DNA的径向延伸以及用于将DNA共价附于蛋白质表面的接头。在50 mM RbCl的实验盐浓度下,Rb +阳离子由于DNA和接头而补偿了约90%的负电荷。高于75 mM时,DFT计算可预测Rb +对DNA电荷的过度补偿。这项研究提出了一种通过预测离子云密度与盐浓度,DNA嫁接密度和长度的函数关系来探索Pro-SNA在不同环境中的结构和功能的方法。全面的,



























 京公网安备 11010802027423号
京公网安备 11010802027423号