当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
PepsNMR for 1 H NMR metabolomic data pre-processing
Analytica Chimica Acta ( IF 6.2 ) Pub Date : 2018-08-01 , DOI: 10.1016/j.aca.2018.02.067 Manon Martin , Benoît Legat , Justine Leenders , Julien Vanwinsberghe , Réjane Rousseau , Bruno Boulanger , Paul H.C. Eilers , Pascal De Tullio , Bernadette Govaerts
Analytica Chimica Acta ( IF 6.2 ) Pub Date : 2018-08-01 , DOI: 10.1016/j.aca.2018.02.067 Manon Martin , Benoît Legat , Justine Leenders , Julien Vanwinsberghe , Réjane Rousseau , Bruno Boulanger , Paul H.C. Eilers , Pascal De Tullio , Bernadette Govaerts
|
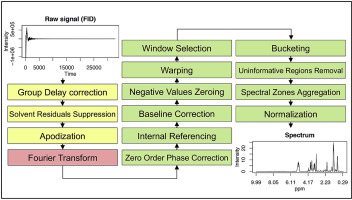
In the analysis of biological samples, control over experimental design and data acquisition procedures alone cannot ensure well-conditioned 1H NMR spectra with maximal information recovery for data analysis. A third major element affects the accuracy and robustness of results: the data pre-processing/pre-treatment for which not enough attention is usually devoted, in particular in metabolomic studies. The usual approach is to use proprietary software provided by the analytical instruments' manufacturers to conduct the entire pre-processing strategy. This widespread practice has a number of advantages such as a user-friendly interface with graphical facilities, but it involves non-negligible drawbacks: a lack of methodological information and automation, a dependency of subjective human choices, only standard processing possibilities and an absence of objective quality criteria to evaluate pre-processing quality. This paper introduces PepsNMR to meet these needs, an R package dedicated to the whole processing chain prior to multivariate data analysis, including, among other tools, solvent signal suppression, internal calibration, phase, baseline and misalignment corrections, bucketing and normalisation. Methodological aspects are discussed and the package is compared to the gold standard procedure with two metabolomic case studies. The use of PepsNMR on these data shows better information recovery and predictive power based on objective and quantitative quality criteria. Other key assets of the package are workflow processing speed, reproducibility, reporting and flexibility, graphical outputs and documented routines.
中文翻译:

PepsNMR 用于 1 H NMR 代谢组学数据预处理
在生物样品的分析中,单独控制实验设计和数据采集程序并不能确保条件良好的 1H NMR 光谱具有最大的数据分析信息恢复。第三个主要因素影响结果的准确性和稳健性:通常没有给予足够重视的数据预处理/预处理,特别是在代谢组学研究中。通常的方法是使用分析仪器制造商提供的专有软件来执行整个预处理策略。这种广泛的做法有许多优点,例如具有图形设施的用户友好界面,但它涉及不可忽视的缺点:缺乏方法信息和自动化,依赖主观的人类选择,只有标准的加工可能性,缺乏客观的质量标准来评估预处理质量。本文介绍了 PepsNMR 来满足这些需求,这是一个 R 包,专用于多变量数据分析之前的整个处理链,其中包括溶剂信号抑制、内部校准、相位、基线和未对准校正、分桶和归一化等工具。讨论了方法学方面,并将该软件包与具有两个代谢组学案例研究的黄金标准程序进行了比较。PepsNMR 对这些数据的使用显示了基于客观和定量质量标准的更好的信息恢复和预测能力。该软件包的其他关键资产是工作流程处理速度、再现性、报告和灵活性、图形输出和记录的例程。
更新日期:2018-08-01
中文翻译:

PepsNMR 用于 1 H NMR 代谢组学数据预处理
在生物样品的分析中,单独控制实验设计和数据采集程序并不能确保条件良好的 1H NMR 光谱具有最大的数据分析信息恢复。第三个主要因素影响结果的准确性和稳健性:通常没有给予足够重视的数据预处理/预处理,特别是在代谢组学研究中。通常的方法是使用分析仪器制造商提供的专有软件来执行整个预处理策略。这种广泛的做法有许多优点,例如具有图形设施的用户友好界面,但它涉及不可忽视的缺点:缺乏方法信息和自动化,依赖主观的人类选择,只有标准的加工可能性,缺乏客观的质量标准来评估预处理质量。本文介绍了 PepsNMR 来满足这些需求,这是一个 R 包,专用于多变量数据分析之前的整个处理链,其中包括溶剂信号抑制、内部校准、相位、基线和未对准校正、分桶和归一化等工具。讨论了方法学方面,并将该软件包与具有两个代谢组学案例研究的黄金标准程序进行了比较。PepsNMR 对这些数据的使用显示了基于客观和定量质量标准的更好的信息恢复和预测能力。该软件包的其他关键资产是工作流程处理速度、再现性、报告和灵活性、图形输出和记录的例程。


























 京公网安备 11010802027423号
京公网安备 11010802027423号