Current Opinion in Chemical Engineering ( IF 6.6 ) Pub Date : 2018-03-09 , DOI: 10.1016/j.coche.2018.02.005 Laurence Calzone , Emmanuel Barillot , Andrei Zinovyev
|
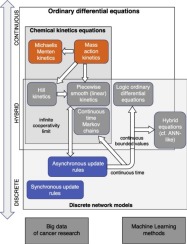
Mathematical modeling of biological networks is a promising approach to understand the complexity of cancer progression, which can be understood as accumulated abnormalities in the kinetics of cellular biochemistry. Two major modeling formalisms (languages) have been used for this purpose in the last couple of decades: one is based on the application of classical chemical kinetics of reaction networks and the other one is based on discrete kinetics representation (called logical formalism for simplicity here), governed by logical state update rules. In this short review, we remind the reader how these two methodologies complement each other but also present the fast and recent development of semi-quantitative approaches for modeling large biological networks, with a spectrum of complementary ideas each inheriting and combining features of both modeling formalisms. We also notice an increasing influence of the recent success of machine learning and artificial intelligence onto the methodology of mathematical network modeling in cancer research, leading to appearance of a number of pragmatic hybrid approaches.
To illustrate the two approaches, logical versus kinetic modeling, we provide an example describing the same biological process with different description granularity in both discrete and continuous formalisms. The model focuses on a central question in cancer biology: understanding the mechanisms of metastasis appearance.
We conclude that despite significant progress in development of modeling ideas, predicting response of large biological networks involved in cancer to various perturbations remains a major unsolved challenge in cancer systems biology.
中文翻译:

生物网络的逻辑模型与动力学模型:在癌症研究中的应用
生物网络的数学建模是一种了解癌症进展复杂性的有前途的方法,可以将其理解为细胞生物化学动力学中累积的异常。在过去的几十年中,已经使用了两种主要的建模形式主义(语言):一种是基于反应网络的经典化学动力学的应用,另一种是基于离散动力学表示(为简单起见,称为逻辑形式主义)。 ),由逻辑状态更新规则控制。在这篇简短的评论中,我们提醒读者这两种方法是如何相互补充的,而且还介绍了用于对大型生物网络进行建模的半定量方法的快速发展和最新进展,具有一系列互补的思想,每个思想都继承并结合了两种建模形式主义的特征。我们还注意到最近机器学习和人工智能的成功对癌症研究中的数学网络建模方法的影响越来越大,导致出现了许多实用的混合方法。
为了说明逻辑建模和动力学建模这两种方法,我们提供了一个示例,以离散形式和连续形式形式描述具有不同描述粒度的相同生物过程。该模型关注癌症生物学中的一个中心问题:了解转移灶出现的机制。
我们得出的结论是,尽管建模思想的发展取得了长足进步,但预测参与癌症的大型生物网络对各种干扰的反应仍然是癌症系统生物学中尚未解决的主要挑战。


























 京公网安备 11010802027423号
京公网安备 11010802027423号