当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Exploring the Potential of Data-Independent Acquisition Proteomics Using Untargeted All-Ion Quantitation: Application to Tumor Subtype Diagnosis
Analytical Chemistry ( IF 7.4 ) Pub Date : 2018-03-09 00:00:00 , DOI: 10.1021/acs.analchem.7b03920 Zhixiang Yan 1, 2 , Ru Yan 1, 2
Analytical Chemistry ( IF 7.4 ) Pub Date : 2018-03-09 00:00:00 , DOI: 10.1021/acs.analchem.7b03920 Zhixiang Yan 1, 2 , Ru Yan 1, 2
Affiliation
|
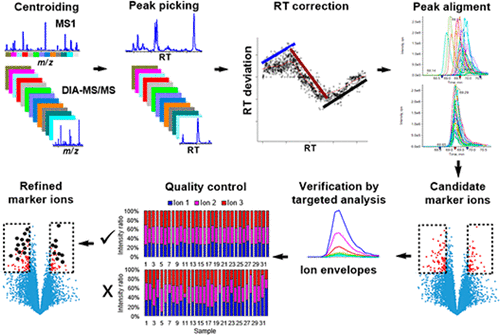
Maximizing the recovery of meaningful biological information can facilitate proteomics-guided early detection and precise treatment of diseases. However, the conventional protein and peptide level targeted quantification of untargeted data independent acquisition (DIA) such as sequential window acquisition of all theoretical spectra (SWATH) is not necessarily descriptive of all information. Untargeted all-ion quantification theoretically could retrieve more features in SWATH digital maps by circumventing the initial identification process but is intrinsically susceptible to errors because of the extreme complexity of proteome samples and the poor selectivity of a single ion. In this study, we optimized and applied the untargeted all-ion quantification of SWATH data to differentiate tumor subtypes. Large peptides and low abundant peptides benefited more from untargeted all-ion quantification. Top-ranked significant ions were linked to their corresponding ion envelops, where multiple correlated ions were used for measurement and only ion envelopes containing at least three ions with consistent intensity ratio were kept as refined differentiating features. Multivariate statistical analysis revealed that for the tested data set, the refined markers discovered by untargeted SWATH analysis showed comparable diagnostic power to protein and peptide markers. Limitations and benefits of the approach are further discussed.
中文翻译:

使用非靶向全离子定量探索数据独立的采集蛋白质组学的潜力:在肿瘤亚型诊断中的应用
最大限度地回收有意义的生物学信息可以促进蛋白质组学指导的疾病的早期发现和精确治疗。但是,常规的蛋白质和肽水平靶向的非目标数据独立采集(DIA),例如所有理论光谱的连续窗口采集(SWATH),不一定能描述所有信息。从理论上讲,无靶全离子定量分析可以通过避开最初的鉴定过程而在SWATH数字地图中检索更多特征,但由于蛋白质组样品的极端复杂性和单个离子的选择性差,因此本质上容易出错。在这项研究中,我们优化并应用了SWATH数据的非目标全离子定量,以区分肿瘤亚型。大肽和低丰度肽得益于非靶向全离子定量分析。排名靠前的重要离子与它们对应的离子包络相关联,其中使用多个相关离子进行测量,只有包含至少三个强度比一致的离子的离子包膜才被保留为精细的区分特征。多变量统计分析表明,对于测试数据集,通过非靶向SWATH分析发现的精制标记显示出与蛋白质和肽标记相当的诊断能力。进一步讨论了该方法的局限性和好处。其中使用多个相关离子进行测量,仅保留包含至少三个强度比一致的离子的离子包膜作为精细的区分特征。多变量统计分析表明,对于测试数据集,通过非靶向SWATH分析发现的精制标记显示出与蛋白质和肽标记相当的诊断能力。进一步讨论了该方法的局限性和好处。其中使用多个相关离子进行测量,仅保留包含至少三个强度比一致的离子的离子包膜作为精细的区分特征。多变量统计分析表明,对于测试数据集,通过非靶向SWATH分析发现的精制标记显示出与蛋白质和肽标记相当的诊断能力。进一步讨论了该方法的局限性和好处。
更新日期:2018-03-09
中文翻译:

使用非靶向全离子定量探索数据独立的采集蛋白质组学的潜力:在肿瘤亚型诊断中的应用
最大限度地回收有意义的生物学信息可以促进蛋白质组学指导的疾病的早期发现和精确治疗。但是,常规的蛋白质和肽水平靶向的非目标数据独立采集(DIA),例如所有理论光谱的连续窗口采集(SWATH),不一定能描述所有信息。从理论上讲,无靶全离子定量分析可以通过避开最初的鉴定过程而在SWATH数字地图中检索更多特征,但由于蛋白质组样品的极端复杂性和单个离子的选择性差,因此本质上容易出错。在这项研究中,我们优化并应用了SWATH数据的非目标全离子定量,以区分肿瘤亚型。大肽和低丰度肽得益于非靶向全离子定量分析。排名靠前的重要离子与它们对应的离子包络相关联,其中使用多个相关离子进行测量,只有包含至少三个强度比一致的离子的离子包膜才被保留为精细的区分特征。多变量统计分析表明,对于测试数据集,通过非靶向SWATH分析发现的精制标记显示出与蛋白质和肽标记相当的诊断能力。进一步讨论了该方法的局限性和好处。其中使用多个相关离子进行测量,仅保留包含至少三个强度比一致的离子的离子包膜作为精细的区分特征。多变量统计分析表明,对于测试数据集,通过非靶向SWATH分析发现的精制标记显示出与蛋白质和肽标记相当的诊断能力。进一步讨论了该方法的局限性和好处。其中使用多个相关离子进行测量,仅保留包含至少三个强度比一致的离子的离子包膜作为精细的区分特征。多变量统计分析表明,对于测试数据集,通过非靶向SWATH分析发现的精制标记显示出与蛋白质和肽标记相当的诊断能力。进一步讨论了该方法的局限性和好处。


























 京公网安备 11010802027423号
京公网安备 11010802027423号