PLoS Pathogens ( IF 6.7 ) Pub Date : 2018-03-05 , DOI: 10.1371/journal.ppat.1006939 Allison F. Carey , Jeremy M. Rock , Inna V. Krieger , Michael R. Chase , Marta Fernandez-Suarez , Sebastien Gagneux , James C. Sacchettini , Thomas R. Ioerger , Sarah M. Fortune
|
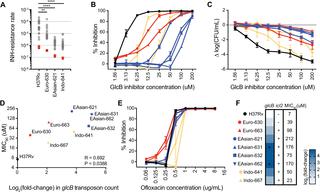
Once considered a phenotypically monomorphic bacterium, there is a growing body of work demonstrating heterogeneity among Mycobacterium tuberculosis (Mtb) strains in clinically relevant characteristics, including virulence and response to antibiotics. However, the genetic and molecular basis for most phenotypic differences among Mtb strains remains unknown. To investigate the basis of strain variation in Mtb, we performed genome-wide transposon mutagenesis coupled with next-generation sequencing (TnSeq) for a panel of Mtb clinical isolates and the reference strain H37Rv to compare genetic requirements for in vitro growth across these strains. We developed an analytic approach to identify quantitative differences in genetic requirements between these genetically diverse strains, which vary in genomic structure and gene content. Using this methodology, we found differences between strains in their requirements for genes involved in fundamental cellular processes, including redox homeostasis and central carbon metabolism. Among the genes with differential requirements were katG, which encodes the activator of the first-line antitubercular agent isoniazid, and glcB, which encodes malate synthase, the target of a novel small-molecule inhibitor. Differences among strains in their requirement for katG and glcB predicted differences in their response to these antimicrobial agents. Importantly, these strain-specific differences in antibiotic response could not be predicted by genetic variants identified through whole genome sequencing or by gene expression analysis. Our results provide novel insight into the basis of variation among Mtb strains and demonstrate that TnSeq is a scalable method to predict clinically important phenotypic differences among Mtb strains.
中文翻译:

结核分枝杆菌临床分离株的TnSeq显示出菌株特异性抗生素的作用
一旦被认为是表型单态细菌,现在越来越多的工作证明结核分枝杆菌(Mtb)菌株在临床相关特征(包括毒力和对抗生素的反应)方面具有异质性。但是,Mtb菌株之间大多数表型差异的遗传和分子基础仍然未知。为了研究Mtb中菌株变异的基础,我们对一组Mtb临床分离株和参考菌株H37Rv进行了全基因组转座子诱变和下一代测序(TnSeq),以比较体外的遗传要求这些菌株的生长。我们开发了一种分析方法来鉴定这些遗传多样性菌株之间遗传需求的数量差异,这些差异在基因组结构和基因含量上有所不同。使用这种方法,我们发现了菌株之间对参与基本细胞过程(包括氧化还原稳态和中央碳代谢)的基因的要求之间存在差异。在具有不同需求的基因中,有编码一线抗结核药物异烟肼激活剂的katG和编码苹果酸合酶(一种新型小分子抑制剂的目标)的glcB。菌株对katG和glcB的需求差异预测了它们对这些抗菌剂的反应差异。重要的是,无法通过通过全基因组测序或基因表达分析鉴定出的遗传变异来预测抗生素反应中这些菌株特异性的差异。我们的结果提供了新的见解,以了解Mtb菌株之间变异的基础,并证明TnSeq是一种可预测的方法,可预测Mtb菌株之间临床上重要的表型差异。


























 京公网安备 11010802027423号
京公网安备 11010802027423号