PLOS Neglected Tropical Diseases ( IF 3.8 ) Pub Date : 2018-03-01 , DOI: 10.1371/journal.pntd.0006254 Dagwin Luang-Suarkia , Oriol Mitja , Timo Ernst , Shannon Bennett , Alfred Tay , Russell Hays , David W. Smith , Allison Imrie
|
Background
Dengue is endemic in the Western Pacific and Oceania and the region reports more than 200,000 cases annually. Outbreaks of dengue and severe dengue occur regularly and movement of virus throughout the region has been reported. Disease surveillance systems, however, in many areas are not fully established and dengue incidence is underreported. Dengue epidemiology is likely least understood in Papua New Guinea (PNG), where the prototype DENV-2 strain New Guinea C was first isolated by Sabin in 1944 but where routine surveillance is not undertaken and little incidence and prevalence data is available.
Methodology/Principal findings
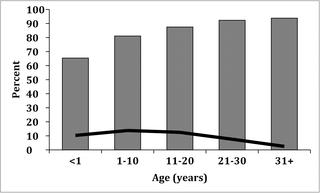
Serum samples from individuals with recent acute febrile illness or with non-febrile conditions collected between 2007–2010 were tested for anti-DENV neutralizing antibody. Responses were predominantly multitypic and seroprevalence increased with age, a pattern indicative of endemic dengue. DENV-1, DENV-2 and DENV-3 genomes were detected by RT-PCR within a nine-month period and in several instances, two serotypes were identified in individuals sampled within a period of 10 days. Phylogenetic analysis of whole genome sequences identified a DENV-3 Genotype 1 lineage which had evolved on the northern coast of PNG which was likely exported to the western Pacific five years later, in addition to a DENV-2 Cosmopolitan Genotype lineage which had previously circulated in the region.
Conclusions/Significance
We show that dengue is hyperendemic in PNG and identify an endemic, locally evolved lineage of DENV-3 that was associated with an outbreak of severe dengue in Pacific countries in subsequent years, although severe disease was not identified in PNG. Additional studies need to be undertaken to understand dengue epidemiology and burden of disease in PNG.
中文翻译:

高流行性登革热的传播和局部进化的DENV-3谱系的鉴定,巴布亚新几内亚2007-2010
背景
登革热是西太平洋和大洋洲的地方病,该地区每年报告超过200,000例病例。登革热和严重登革热的暴发经常发生,据报道病毒在整个地区传播。但是,在许多地区,疾病监测系统尚未完全建立,登革热发病率报道不足。登革热流行病学在巴布亚新几内亚(PNG)中了解得最少,那里的原型DENV-2株新几内亚C最初是由萨宾(Sabin)在1944年分离出来的,但当时没有进行常规监测,而且几乎没有发病率和患病率数据。
方法/主要发现
测试了2007-2010年之间近期患有急性发热性疾病或非发热性疾病患者的血清样本中的抗DENV中和抗体。反应主要是多型的,血清阳性率随年龄增加而增加,这是登革热的一种流行趋势。通过RT-PCR在9个月内检测到DENV-1,DENV-2和DENV-3基因组,在几种情况下,在10天内取样的个体中鉴定出两种血清型。对整个基因组序列进行系统进化分析,确定了DENV-3基因型1谱系,该谱系已在PNG北部海岸演化,五年后可能已出口到西太平洋,此外,先前已在DENV-2世界性基因型谱系中传播。该区域。
结论/意义
我们显示登革热在巴布亚新几内亚是高流行性的,并且确定了随后几年与太平洋国家中严重登革热暴发有关的地方性,局部进化的DENV-3谱系,尽管在巴布亚新几内亚未发现严重的疾病。需要进行其他研究来了解登革热的登革热流行病学和疾病负担。



























 京公网安备 11010802027423号
京公网安备 11010802027423号