Current Opinion in Chemical Engineering ( IF 6.6 ) Pub Date : 2018-01-11 , DOI: 10.1016/j.coche.2017.12.001 Michael L Paull , Patrick S Daugherty
|
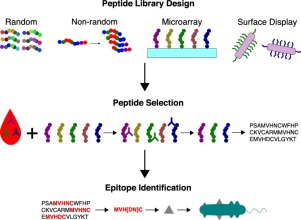
Antibodies in blood provide a rich source of immunological information. Antibody repertoire analysis seeks to decode this information to empower the development of vaccines, diagnostics, and therapeutics. To this end, various approaches have been developed to determine epitopes using peptide libraries. Approaches have used random or proteome-derived peptide libraries in a microarray or surface display format. For methods using random libraries, motif discovery software has been developed to identify common binding signatures. The analysis of thousands of samples and dozens of diseases has shown that there are often disease-specific epitopes, even though individual antibody repertoires are unique. The recent developments in antibody repertoire analysis hold the potential to enable comprehensive immune evaluations.
中文翻译:

使用肽库定位血清抗体库
血液中的抗体提供了丰富的免疫学信息。抗体库分析试图对这些信息进行解码,以授权开发疫苗,诊断和治疗方法。为此,已经开发了使用肽文库确定表位的各种方法。方法已经以微阵列或表面展示形式使用了随机的或蛋白质组衍生的肽库。对于使用随机库的方法,已开发出基序发现软件来识别常见的结合签名。对成千上万个样本和数十种疾病的分析表明,尽管单个抗体库是唯一的,但通常也存在特定于疾病的表位。抗体库分析的最新发展具有进行全面免疫评估的潜力。



























 京公网安备 11010802027423号
京公网安备 11010802027423号