当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Binding Modes of Ligands Using Enhanced Sampling (BLUES): Rapid Decorrelation of Ligand Binding Modes via Nonequilibrium Candidate Monte Carlo
The Journal of Physical Chemistry B ( IF 3.3 ) Pub Date : 2018-02-27 00:00:00 , DOI: 10.1021/acs.jpcb.7b11820 Samuel C. Gill 1 , Nathan M. Lim 2 , Patrick B. Grinaway 3, 4 , Ariën S. Rustenburg 3, 4 , Josh Fass 4, 5 , Gregory A. Ross 4 , John D. Chodera 6 , David L. Mobley 1, 2
The Journal of Physical Chemistry B ( IF 3.3 ) Pub Date : 2018-02-27 00:00:00 , DOI: 10.1021/acs.jpcb.7b11820 Samuel C. Gill 1 , Nathan M. Lim 2 , Patrick B. Grinaway 3, 4 , Ariën S. Rustenburg 3, 4 , Josh Fass 4, 5 , Gregory A. Ross 4 , John D. Chodera 6 , David L. Mobley 1, 2
Affiliation
|
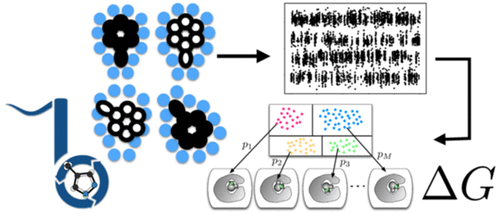
Accurately predicting protein–ligand binding affinities and binding modes is a major goal in computational chemistry, but even the prediction of ligand binding modes in proteins poses major challenges. Here, we focus on solving the binding mode prediction problem for rigid fragments. That is, we focus on computing the dominant placement, conformation, and orientations of a relatively rigid, fragment-like ligand in a receptor, and the populations of the multiple binding modes which may be relevant. This problem is important in its own right, but is even more timely given the recent success of alchemical free energy calculations. Alchemical calculations are increasingly used to predict binding free energies of ligands to receptors. However, the accuracy of these calculations is dependent on proper sampling of the relevant ligand binding modes. Unfortunately, ligand binding modes may often be uncertain, hard to predict, and/or slow to interconvert on simulation time scales, so proper sampling with current techniques can require prohibitively long simulations. We need new methods which dramatically improve sampling of ligand binding modes. Here, we develop and apply a nonequilibrium candidate Monte Carlo (NCMC) method to improve sampling of ligand binding modes. In this technique, the ligand is rotated and subsequently allowed to relax in its new position through alchemical perturbation before accepting or rejecting the rotation and relaxation as a nonequilibrium Monte Carlo move. When applied to a T4 lysozyme model binding system, this NCMC method shows over 2 orders of magnitude improvement in binding mode sampling efficiency compared to a brute force molecular dynamics simulation. This is a first step toward applying this methodology to pharmaceutically relevant binding of fragments and, eventually, drug-like molecules. We are making this approach available via our new Binding modes of ligands using enhanced sampling (BLUES) package which is freely available on GitHub.
中文翻译:

使用增强采样(BLUES)的配体结合模式:通过非平衡候选蒙特卡洛快速解配体结合模式
准确预测蛋白质与配体的结合亲和力和结合方式是计算化学的主要目标,但是即使预测蛋白质中的配体结合方式也构成了重大挑战。在这里,我们专注于解决刚性碎片的结合模式预测问题。也就是说,我们专注于计算受体中相对刚性,片段状配体的主要位置,构象和方向,以及可能相关的多种结合模式。这个问题本身很重要,但是鉴于最近炼金术自由能计算的成功,这个问题显得更为及时。炼金术的计算越来越多地用于预测配体与受体的结合自由能。但是,这些计算的准确性取决于相关配体结合模式的正确采样。不幸的是,配体结合模式通常可能不确定,难以预测和/或在模拟时标上互变速度慢,因此使用当前技术进行正确的采样可能会要求进行冗长的模拟。我们需要能够显着改善配体结合模式采样的新方法。在这里,我们开发并应用非平衡候选蒙特卡洛(NCMC)方法来改善配体结合模式的采样。在这种技术中,配体旋转,随后通过炼金术扰动使其松弛到新的位置,然后接受或拒绝旋转和松弛作为非平衡蒙特卡洛运动。当应用于T4溶菌酶模型结合系统时,与蛮力分子动力学模拟相比,这种NCMC方法显示结合模式采样效率提高了2个数量级。这是将该方法应用于片段以及最终与药物样分子的药物相关结合的第一步。我们正在使用增强采样(BLUES)包通过配体的新绑定模式通过GitHub免费提供此方法。
更新日期:2018-02-27
中文翻译:

使用增强采样(BLUES)的配体结合模式:通过非平衡候选蒙特卡洛快速解配体结合模式
准确预测蛋白质与配体的结合亲和力和结合方式是计算化学的主要目标,但是即使预测蛋白质中的配体结合方式也构成了重大挑战。在这里,我们专注于解决刚性碎片的结合模式预测问题。也就是说,我们专注于计算受体中相对刚性,片段状配体的主要位置,构象和方向,以及可能相关的多种结合模式。这个问题本身很重要,但是鉴于最近炼金术自由能计算的成功,这个问题显得更为及时。炼金术的计算越来越多地用于预测配体与受体的结合自由能。但是,这些计算的准确性取决于相关配体结合模式的正确采样。不幸的是,配体结合模式通常可能不确定,难以预测和/或在模拟时标上互变速度慢,因此使用当前技术进行正确的采样可能会要求进行冗长的模拟。我们需要能够显着改善配体结合模式采样的新方法。在这里,我们开发并应用非平衡候选蒙特卡洛(NCMC)方法来改善配体结合模式的采样。在这种技术中,配体旋转,随后通过炼金术扰动使其松弛到新的位置,然后接受或拒绝旋转和松弛作为非平衡蒙特卡洛运动。当应用于T4溶菌酶模型结合系统时,与蛮力分子动力学模拟相比,这种NCMC方法显示结合模式采样效率提高了2个数量级。这是将该方法应用于片段以及最终与药物样分子的药物相关结合的第一步。我们正在使用增强采样(BLUES)包通过配体的新绑定模式通过GitHub免费提供此方法。


























 京公网安备 11010802027423号
京公网安备 11010802027423号