当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Theory of Sequence Effects in Amyloid Aggregation
The Journal of Physical Chemistry B ( IF 3.3 ) Pub Date : 2018-02-27 00:00:00 , DOI: 10.1021/acs.jpcb.7b11830 Caleb Huang 1 , Elaheh Ghanati 1 , Jeremy D. Schmit 1
The Journal of Physical Chemistry B ( IF 3.3 ) Pub Date : 2018-02-27 00:00:00 , DOI: 10.1021/acs.jpcb.7b11830 Caleb Huang 1 , Elaheh Ghanati 1 , Jeremy D. Schmit 1
Affiliation
|
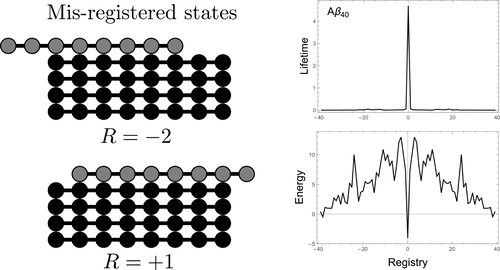
We present a simple model for the effect of amino acid sequences on amyloid fibril formation. Using the HP model we find the binding lifetimes of four simple sequences by solving the first passage time for the intermolecular H-bond reaction coordinate. We find that sequences with identical binding energies have widely varying binding times depending on where the aggregation prone amino acids are located in the sequence. In general, longer binding times occur when the aggregation prone amino acids are clustered in a single “hot spot”. Similarly, binding times are shortened by clustering weakly bound residues. Both of these effects are explained by an increase in the multiplicity of unbinding trajectories that comes from adding weak binding residues. Our model predicts a transition from ordered to disordered fibrils as the concentration of monomers increases. We apply our model to Aβ, IAPP, and apomyoglobin using binding energy estimates derived from bioinformatics. We find that these sequences are highly selective of the in-register state. This selectivity arises from the having strongly bound segments of varying length and separation.
中文翻译:

淀粉样蛋白聚集的序列效应理论
我们提出了一个简单的模型,对淀粉样蛋白原纤维形成氨基酸序列的影响。使用HP模型,我们通过求解分子间H键反应坐标的第一次通过时间来找到四个简单序列的结合寿命。我们发现具有相同结合能的序列具有广泛变化的结合时间,这取决于倾向于聚合的氨基酸在序列中的位置。通常,当倾向于聚合的氨基酸聚集在单个“热点”中时,结合时间会更长。类似地,通过将弱结合的残基聚类来缩短结合时间。通过添加弱的结合残基,解除结合轨迹的多样性的增加可以解释这两种效应。我们的模型预测,随着单体浓度的增加,将从有序的原纤维过渡到无序的原纤维。我们使用源自生物信息学的结合能估计将模型应用到Aβ,IAPP和apomyoglobin。我们发现这些序列对寄存器内状态具有高度选择性。这种选择性源自具有可变长度和间隔的牢固结合的链段。
更新日期:2018-02-27
中文翻译:

淀粉样蛋白聚集的序列效应理论
我们提出了一个简单的模型,对淀粉样蛋白原纤维形成氨基酸序列的影响。使用HP模型,我们通过求解分子间H键反应坐标的第一次通过时间来找到四个简单序列的结合寿命。我们发现具有相同结合能的序列具有广泛变化的结合时间,这取决于倾向于聚合的氨基酸在序列中的位置。通常,当倾向于聚合的氨基酸聚集在单个“热点”中时,结合时间会更长。类似地,通过将弱结合的残基聚类来缩短结合时间。通过添加弱的结合残基,解除结合轨迹的多样性的增加可以解释这两种效应。我们的模型预测,随着单体浓度的增加,将从有序的原纤维过渡到无序的原纤维。我们使用源自生物信息学的结合能估计将模型应用到Aβ,IAPP和apomyoglobin。我们发现这些序列对寄存器内状态具有高度选择性。这种选择性源自具有可变长度和间隔的牢固结合的链段。



























 京公网安备 11010802027423号
京公网安备 11010802027423号