当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Probing Structural Changes among Analogous Inhibitor-Bound Forms of HIV-1 Protease and a Drug-Resistant Mutant in Solution by Nuclear Magnetic Resonance
Biochemistry ( IF 2.9 ) Pub Date : 2018-02-19 00:00:00 , DOI: 10.1021/acs.biochem.7b01238 Shahid N. Khan 1 , John D. Persons 1 , Janet L. Paulsen 2 , Michel Guerrero 1 , Celia A. Schiffer 2 , Nese Kurt-Yilmaz 2 , Rieko Ishima 1
Biochemistry ( IF 2.9 ) Pub Date : 2018-02-19 00:00:00 , DOI: 10.1021/acs.biochem.7b01238 Shahid N. Khan 1 , John D. Persons 1 , Janet L. Paulsen 2 , Michel Guerrero 1 , Celia A. Schiffer 2 , Nese Kurt-Yilmaz 2 , Rieko Ishima 1
Affiliation
|
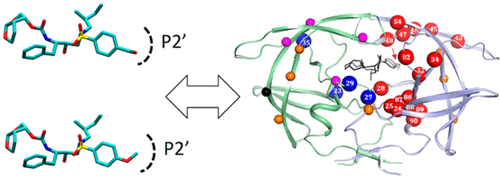
In the era of state-of-the-art inhibitor design and high-resolution structural studies, detection of significant but small protein structural differences in the inhibitor-bound forms is critical to further developing the inhibitor. Here, we probed differences in HIV-1 protease (PR) conformation among darunavir and four analogous inhibitor-bound forms and compared them with a drug-resistant mutant using nuclear magnetic resonance chemical shifts. Changes in amide chemical shifts of wild-type (WT) PR among these inhibitor-bound forms, ΔCSP, were subtle but detectable and extended >10 Å from the inhibitor-binding site, asymmetrically between the two subunits of PR. Molecular dynamics simulations revealed differential local hydrogen bonding as the molecular basis of this remote asymmetric change. Inhibitor-bound forms of the drug-resistant mutant also showed a similar long-range ΔCSP pattern. Differences in ΔCSP values of the WT and the mutant (ΔΔCSPs) were observed at the inhibitor-binding site and in the surrounding region. Comparing chemical shift changes among highly analogous inhibitors and ΔΔCSPs effectively eliminated local environmental effects stemming from different chemical groups and enabled exploitation of these sensitive parameters to detect subtle protein conformational changes and to elucidate asymmetric and remote conformational effects upon inhibitor interaction.
中文翻译:

通过核磁共振探测溶液中HIV-1蛋白酶的类似抑制剂结合形式和抗药性突变体之间的结构变化
在最先进的抑制剂设计和高分辨率结构研究时代,以抑制剂结合形式检测显着但很小的蛋白质结构差异对于进一步开发抑制剂至关重要。在这里,我们探讨了地那那韦和四种类似的抑制剂结合形式在HIV-1蛋白酶(PR)构象上的差异,并将它们与使用核磁共振化学位移的抗药性突变体进行了比较。在这些抑制剂结合形式ΔCSP中,野生型(WT)PR的酰胺化学位移变化很小,但可检测到,并且从抑制剂结合位点在PR的两个亚基之间不对称延伸了10Å以上。分子动力学模拟表明,不同的局部氢键是这种远程不对称变化的分子基础。耐药突变体的抑制剂结合形式也显示出相似的远程ΔCSP模式。在抑制剂结合位点和周围区域观察到WT和突变体的ΔCSP值差异(ΔΔCSPs)。比较高度相似的抑制剂和ΔΔCSP之间的化学位移变化,可以有效地消除源自不同化学基团的局部环境影响,并能够利用这些敏感参数检测微妙的蛋白质构象变化,并阐明抑制剂相互作用时的不对称和远程构象效应。
更新日期:2018-02-19
中文翻译:

通过核磁共振探测溶液中HIV-1蛋白酶的类似抑制剂结合形式和抗药性突变体之间的结构变化
在最先进的抑制剂设计和高分辨率结构研究时代,以抑制剂结合形式检测显着但很小的蛋白质结构差异对于进一步开发抑制剂至关重要。在这里,我们探讨了地那那韦和四种类似的抑制剂结合形式在HIV-1蛋白酶(PR)构象上的差异,并将它们与使用核磁共振化学位移的抗药性突变体进行了比较。在这些抑制剂结合形式ΔCSP中,野生型(WT)PR的酰胺化学位移变化很小,但可检测到,并且从抑制剂结合位点在PR的两个亚基之间不对称延伸了10Å以上。分子动力学模拟表明,不同的局部氢键是这种远程不对称变化的分子基础。耐药突变体的抑制剂结合形式也显示出相似的远程ΔCSP模式。在抑制剂结合位点和周围区域观察到WT和突变体的ΔCSP值差异(ΔΔCSPs)。比较高度相似的抑制剂和ΔΔCSP之间的化学位移变化,可以有效地消除源自不同化学基团的局部环境影响,并能够利用这些敏感参数检测微妙的蛋白质构象变化,并阐明抑制剂相互作用时的不对称和远程构象效应。


























 京公网安备 11010802027423号
京公网安备 11010802027423号