当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Orthogonal Information Encoding in Living Cells with High Error-Tolerance, Safety, and Fidelity
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-02-10 00:00:00 , DOI: 10.1021/acssynbio.7b00382 Lifu Song 1 , An-Ping Zeng 1
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-02-10 00:00:00 , DOI: 10.1021/acssynbio.7b00382 Lifu Song 1 , An-Ping Zeng 1
Affiliation
|
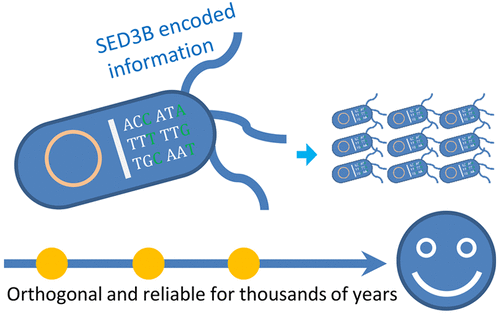
Information encoding in DNA is of great interest but its applications in vivo might be questionable since errors could be enriched exponentially by cellular replications and the artificial sequences may interfere with the natural ones. Here, a novel self-error-detecting, three-base block encoding scheme (SED3B) is proposed for reliable and orthogonal information encoding in living cells. SED3B utilizes a novel way to add error detecting bases in small data blocks which can combine with the inherent redundancy of DNA molecules for effective error correction. Errors at a rate of 19% can be corrected as shown by error-prone PCR experiments with E. coli cells. Calculations based on this preliminary result show that SED3B encoded information in E. coli can be reliable for more than 12 000 years of continuous replication. Importantly, SED3B encoded sequences do not share sequence space to all reported natural DNA sequences except for some short tandem repeats, indicating a low biological relevance of encoded sequences for the first time. These features make SED3B attractive for broad orthogonal information encoding purposes in living cells, for example, comments/barcodes encoding in synthetic biology. For proof of concept, 10 different barcodes were encoded in E. coli cells. After continuous replications for 10 days including exposure to ultraviolet for 2–3 min (lethality >60%) per day, all barcodes were fully recovered, proving the stability of the encoded information. An online encoding-decoding system is implemented and available at http://biosystem.bt1.tu-harburg.de/sed3b/.
中文翻译:

具有高容错性,安全性和保真度的活细胞中的正交信息编码
DNA中的信息编码备受关注,但其在体内的应用可能会受到质疑,因为错误可能会因细胞复制而呈指数增加,并且人工序列可能会干扰天然序列。在这里,提出了一种新颖的自检错三基块编码方案(SED3B),用于在活细胞中进行可靠的正交信息编码。SED3B利用一种新颖的方法在小的数据块中添加了错误检测基础,可以将其与DNA分子的固有冗余相结合,以进行有效的错误校正。如对大肠杆菌细胞的易错PCR实验所示,可以纠正19%的错误。根据此初步结果进行的计算表明,SED3B在大肠杆菌中编码信息可以持续超过12000年的持续复制。重要的是,SED3B编码的序列除某些短串联重复序列外,不与所有报告的天然DNA序列共享序列空间,这首次表明编码序列的生物学相关性较低。这些功能使SED3B具有很强的吸引力,可用于活细胞中广泛的正交信息编码,例如合成生物学中的注释/条形码编码。为了证明概念,在大肠杆菌中编码了10种不同的条形码细胞。连续复制10天(包括每天暴露于紫外线2至3分钟(致死率> 60%))后,所有条形码均已完全回收,证明了编码信息的稳定性。实现了一种在线编码解码系统,该系统可从http://biosystem.bt1.tu-harburg.de/sed3b/获得。
更新日期:2018-02-10
中文翻译:

具有高容错性,安全性和保真度的活细胞中的正交信息编码
DNA中的信息编码备受关注,但其在体内的应用可能会受到质疑,因为错误可能会因细胞复制而呈指数增加,并且人工序列可能会干扰天然序列。在这里,提出了一种新颖的自检错三基块编码方案(SED3B),用于在活细胞中进行可靠的正交信息编码。SED3B利用一种新颖的方法在小的数据块中添加了错误检测基础,可以将其与DNA分子的固有冗余相结合,以进行有效的错误校正。如对大肠杆菌细胞的易错PCR实验所示,可以纠正19%的错误。根据此初步结果进行的计算表明,SED3B在大肠杆菌中编码信息可以持续超过12000年的持续复制。重要的是,SED3B编码的序列除某些短串联重复序列外,不与所有报告的天然DNA序列共享序列空间,这首次表明编码序列的生物学相关性较低。这些功能使SED3B具有很强的吸引力,可用于活细胞中广泛的正交信息编码,例如合成生物学中的注释/条形码编码。为了证明概念,在大肠杆菌中编码了10种不同的条形码细胞。连续复制10天(包括每天暴露于紫外线2至3分钟(致死率> 60%))后,所有条形码均已完全回收,证明了编码信息的稳定性。实现了一种在线编码解码系统,该系统可从http://biosystem.bt1.tu-harburg.de/sed3b/获得。



























 京公网安备 11010802027423号
京公网安备 11010802027423号