当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Recurrent Neural Network Model for Constructive Peptide Design
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-22 00:00:00 , DOI: 10.1021/acs.jcim.7b00414 Alex T. Müller 1 , Jan A. Hiss 1 , Gisbert Schneider 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-22 00:00:00 , DOI: 10.1021/acs.jcim.7b00414 Alex T. Müller 1 , Jan A. Hiss 1 , Gisbert Schneider 1
Affiliation
|
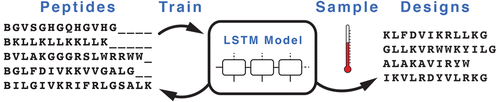
We present a generative long short-term memory (LSTM) recurrent neural network (RNN) for combinatorial de novo peptide design. RNN models capture patterns in sequential data and generate new data instances from the learned context. Amino acid sequences represent a suitable input for these machine-learning models. Generative models trained on peptide sequences could therefore facilitate the design of bespoke peptide libraries. We trained RNNs with LSTM units on pattern recognition of helical antimicrobial peptides and used the resulting model for de novo sequence generation. Of these sequences, 82% were predicted to be active antimicrobial peptides compared to 65% of randomly sampled sequences with the same amino acid distribution as the training set. The generated sequences also lie closer to the training data than manually designed amphipathic helices. The results of this study showcase the ability of LSTM RNNs to construct new amino acid sequences within the applicability domain of the model and motivate their prospective application to peptide and protein design without the need for the exhaustive enumeration of sequence libraries.
中文翻译:

递归神经网络模型的建设性肽设计。
我们提出了用于组合从头肽设计的生成性长短期记忆(LSTM)递归神经网络(RNN)。RNN模型捕获顺序数据中的模式,并从学习的上下文中生成新的数据实例。氨基酸序列代表了这些机器学习模型的合适输入。因此,在肽序列上训练的生成模型可以促进定制肽库的设计。我们使用LSTM单元对RNN进行了螺旋抗菌肽的模式识别训练,并将所得模型用于从头序列生成。在这些序列中,预计有82%是有活性的抗菌肽,相比之下,随机抽样的序列中有65%具有与训练集相同的氨基酸分布。生成的序列也比人工设计的两亲螺旋更接近训练数据。这项研究的结果展示了LSTM RNN在模型的适用范围内构建新氨基酸序列的能力,并激发了它们在肽和蛋白质设计中的前瞻性应用,而无需详尽列举序列库。
更新日期:2018-01-22
中文翻译:

递归神经网络模型的建设性肽设计。
我们提出了用于组合从头肽设计的生成性长短期记忆(LSTM)递归神经网络(RNN)。RNN模型捕获顺序数据中的模式,并从学习的上下文中生成新的数据实例。氨基酸序列代表了这些机器学习模型的合适输入。因此,在肽序列上训练的生成模型可以促进定制肽库的设计。我们使用LSTM单元对RNN进行了螺旋抗菌肽的模式识别训练,并将所得模型用于从头序列生成。在这些序列中,预计有82%是有活性的抗菌肽,相比之下,随机抽样的序列中有65%具有与训练集相同的氨基酸分布。生成的序列也比人工设计的两亲螺旋更接近训练数据。这项研究的结果展示了LSTM RNN在模型的适用范围内构建新氨基酸序列的能力,并激发了它们在肽和蛋白质设计中的前瞻性应用,而无需详尽列举序列库。


























 京公网安备 11010802027423号
京公网安备 11010802027423号