当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Improved Descriptors for the Quantitative Structure–Activity Relationship Modeling of Peptides and Proteins
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-16 00:00:00 , DOI: 10.1021/acs.jcim.7b00488 Mark H Barley 1 , Nicholas J Turner 1 , Royston Goodacre 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-16 00:00:00 , DOI: 10.1021/acs.jcim.7b00488 Mark H Barley 1 , Nicholas J Turner 1 , Royston Goodacre 1
Affiliation
|
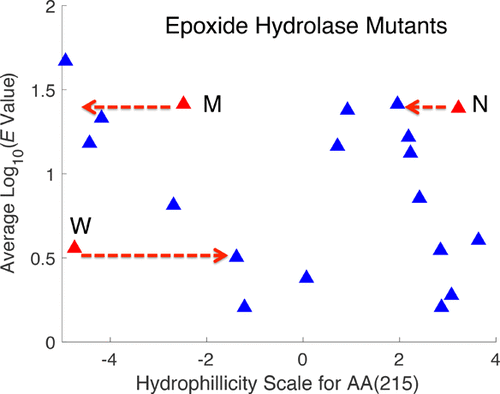
The ability to model the activity of a protein using quantitative structure–activity relationships (QSAR) requires descriptors for the 20 naturally coded amino acids. In this work we show that by modifying some established descriptors we were able to model the activity data of 140 mutants of the enzyme epoxide hydrolase with improved accuracy. These new descriptors (referred to as physical descriptors) also gave very good results when tested against a series of four dipeptide data sets. The physical descriptors encode the amino acids using only two orthogonal scales: the first is strongly linked to hydrophilicity/hydrophobicity, and the second, to the volume of the amino acid residue. The use of these new amino acid descriptors should result in simpler and more readily interpretable models for the enzyme activity (and potentially other functions of interest, e.g., secondary and tertiary structure) of peptides and proteins.
中文翻译:

肽和蛋白质定量结构-活性关系建模的改进描述符
使用定量构效关系 (QSAR) 模拟蛋白质活性的能力需要 20 种天然编码氨基酸的描述符。在这项工作中,我们表明,通过修改一些已建立的描述符,我们能够以更高的准确性对 140 个环氧化物水解酶突变体的活性数据进行建模。这些新的描述符(称为物理描述符)在针对一系列四个二肽数据集进行测试时也给出了非常好的结果。物理描述符仅使用两个正交尺度编码氨基酸:第一个与亲水性/疏水性密切相关,第二个与氨基酸残基的体积相关。
更新日期:2018-01-16
中文翻译:

肽和蛋白质定量结构-活性关系建模的改进描述符
使用定量构效关系 (QSAR) 模拟蛋白质活性的能力需要 20 种天然编码氨基酸的描述符。在这项工作中,我们表明,通过修改一些已建立的描述符,我们能够以更高的准确性对 140 个环氧化物水解酶突变体的活性数据进行建模。这些新的描述符(称为物理描述符)在针对一系列四个二肽数据集进行测试时也给出了非常好的结果。物理描述符仅使用两个正交尺度编码氨基酸:第一个与亲水性/疏水性密切相关,第二个与氨基酸残基的体积相关。



























 京公网安备 11010802027423号
京公网安备 11010802027423号