当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Reconstruction of HMBC Correlation Networks: A Novel NMR-Based Contribution to Metabolite Mixture Analysis
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-31 00:00:00 , DOI: 10.1021/acs.jcim.7b00653 Ali Bakiri 1, 2 , Jane Hubert 1 , Romain Reynaud 2 , Carole Lambert 2 , Agathe Martinez 1 , Jean-Hugues Renault 1 , Jean-Marc Nuzillard 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-31 00:00:00 , DOI: 10.1021/acs.jcim.7b00653 Ali Bakiri 1, 2 , Jane Hubert 1 , Romain Reynaud 2 , Carole Lambert 2 , Agathe Martinez 1 , Jean-Hugues Renault 1 , Jean-Marc Nuzillard 1
Affiliation
|
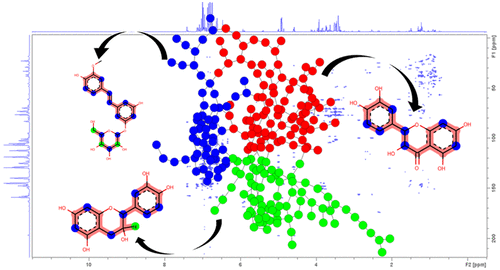
A new in silico method is introduced for the dereplication of natural metabolite mixtures based on HMBC and HSQC spectra that inform about short-range and long-range H–C correlations occurring in the carbon skeleton of individual chemical entities. Starting from the HMBC spectrum of a metabolite mixture, an algorithm was developed in order to recover individualized HMBC footprints of the mixture constituents. The collected H–C correlations are represented by a network of NMR peaks connected to each other when sharing either a 1H or 13C chemical shift value. The network obtained is then divided into clusters using a community detection algorithm, and finally each cluster is tentatively assigned to a molecular structure by means of a NMR chemical shift database containing the theoretical HMBC and HSQC correlation data of a range of natural metabolites. The proof of principle of this method is demonstrated on a model mixture of 3 known natural compounds and then on a real-life bark extract obtained from the common spruce (Picea abies L.).
中文翻译:

HMBC相关网络的重建:基于新的基于NMR的代谢物混合物分析贡献
引入了一种新的计算机模拟方法,用于基于HMBC和HSQC光谱图谱的自然代谢物混合物的重复计算,该图谱揭示了各个化学实体碳骨架中发生的短程和长程H–C相关性。从代谢物混合物的HMBC光谱开始,开发了一种算法,以恢复混合物成分的个性化HMBC足迹。当共享1 H或13 H时,所收集的H–C相关性由相互连接的NMR峰网络表示C化学位移值。然后使用社区检测算法将获得的网络划分为多个簇,最后,通过一个NMR化学位移数据库,将每个簇暂时分配给分子结构,该数据库包含一系列天然代谢物的理论HMBC和HSQC相关数据。在3种已知天然化合物的模型混合物上,然后在从常见云杉(Picea abies L.)获得的真实树皮提取物中,证明了该方法的原理。
更新日期:2018-01-31
中文翻译:

HMBC相关网络的重建:基于新的基于NMR的代谢物混合物分析贡献
引入了一种新的计算机模拟方法,用于基于HMBC和HSQC光谱图谱的自然代谢物混合物的重复计算,该图谱揭示了各个化学实体碳骨架中发生的短程和长程H–C相关性。从代谢物混合物的HMBC光谱开始,开发了一种算法,以恢复混合物成分的个性化HMBC足迹。当共享1 H或13 H时,所收集的H–C相关性由相互连接的NMR峰网络表示C化学位移值。然后使用社区检测算法将获得的网络划分为多个簇,最后,通过一个NMR化学位移数据库,将每个簇暂时分配给分子结构,该数据库包含一系列天然代谢物的理论HMBC和HSQC相关数据。在3种已知天然化合物的模型混合物上,然后在从常见云杉(Picea abies L.)获得的真实树皮提取物中,证明了该方法的原理。



























 京公网安备 11010802027423号
京公网安备 11010802027423号