当前位置:
X-MOL 学术
›
Integr. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Landscape of gene networks for random parameter perturbation
Integrative Biology ( IF 2.5 ) Pub Date : 2018-01-17 , DOI: 10.1039/c7ib00198c Chunhe Li 1, 2, 3, 4, 5
Integrative Biology ( IF 2.5 ) Pub Date : 2018-01-17 , DOI: 10.1039/c7ib00198c Chunhe Li 1, 2, 3, 4, 5
Affiliation
|
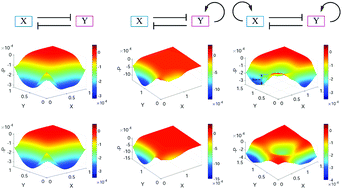
Landscape approaches have been exploited to study the stochastic dynamics of gene networks. However, how to calculate the landscape with a wide range of parameter variations and how to investigate the influence of the network topology on the global properties of gene networks remain to be elucidated. Here, I developed an approach for the landscape of random parameter perturbation (LRPP) to address this issue. Based on a self-consistent approximation approach, by making perturbations to parameters in a given range, I obtained the landscape for gene network systems. I applied this approach to two biological models, one for the mutual repression model and the other for the embryonic stem (ES) cell differentiation network. For the mutual repression model, my results confirm quantitatively that positive feedback promotes the robustness of multistability. For the ES cell differentiation model, I identify three cell states, representing the ES cell, the differentiation cell, and the intermediate state cell, respectively. I propose that the intermediate states and the wide range of parameter values coming from inhomogeneous cellular environments provide possible explanations for the heterogeneity observed in single cell experiments. I also offer a counterintuitive result that noise could reduce heterogeneity and promote the stability of cell states. These results support that the network topology determines the operating principles of the genetic networks, reflected by the representative landscapes from LRPP. This work provides a new route to obtain the potential landscape for a gene network system given a wide range of parameter values and study the influences of the network topology on the global properties of the system.
中文翻译:

基因网络的随机参数摄动
已经利用景观方法来研究基因网络的随机动力学。但是,如何计算具有广泛参数变化的景观以及如何研究网络拓扑结构对基因网络全局特性的影响仍有待阐明。在这里,我针对随机参数摄动(LRPP)的情况开发了一种方法来解决此问题。基于自洽近似方法,通过对给定范围内的参数进行摄动,我获得了基因网络系统的前景。我将这种方法应用于两种生物学模型,一种用于相互抑制模型,另一种用于胚胎干(ES)细胞分化网络。对于互压模型,我的结果从数量上证实了积极的反馈促进了多稳定性的鲁棒性。对于ES细胞分化模型,我确定了三种细胞状态,分别代表ES细胞,分化细胞和中间状态细胞。我建议,来自非均质细胞环境的中间状态和广泛的参数值为单细胞实验中观察到的异质性提供了可能的解释。我还提供了一个违反直觉的结果,即噪声可能会减少异质性并促进细胞状态的稳定性。这些结果表明,网络拓扑结构决定了遗传网络的运行原理,这体现在LRPP的代表性景观上。
更新日期:2018-01-17
中文翻译:

基因网络的随机参数摄动
已经利用景观方法来研究基因网络的随机动力学。但是,如何计算具有广泛参数变化的景观以及如何研究网络拓扑结构对基因网络全局特性的影响仍有待阐明。在这里,我针对随机参数摄动(LRPP)的情况开发了一种方法来解决此问题。基于自洽近似方法,通过对给定范围内的参数进行摄动,我获得了基因网络系统的前景。我将这种方法应用于两种生物学模型,一种用于相互抑制模型,另一种用于胚胎干(ES)细胞分化网络。对于互压模型,我的结果从数量上证实了积极的反馈促进了多稳定性的鲁棒性。对于ES细胞分化模型,我确定了三种细胞状态,分别代表ES细胞,分化细胞和中间状态细胞。我建议,来自非均质细胞环境的中间状态和广泛的参数值为单细胞实验中观察到的异质性提供了可能的解释。我还提供了一个违反直觉的结果,即噪声可能会减少异质性并促进细胞状态的稳定性。这些结果表明,网络拓扑结构决定了遗传网络的运行原理,这体现在LRPP的代表性景观上。



























 京公网安备 11010802027423号
京公网安备 11010802027423号