当前位置:
X-MOL 学术
›
J. Phys. Chem. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Molecular Dynamics Simulations Reveal an Interplay between SHAPE Reagent Binding and RNA Flexibility.
The Journal of Physical Chemistry Letters ( IF 5.7 ) Pub Date : 2018-01-04 , DOI: 10.1021/acs.jpclett.7b02921 Vojtěch Mlýnský 1 , Giovanni Bussi 1
The Journal of Physical Chemistry Letters ( IF 5.7 ) Pub Date : 2018-01-04 , DOI: 10.1021/acs.jpclett.7b02921 Vojtěch Mlýnský 1 , Giovanni Bussi 1
Affiliation
|
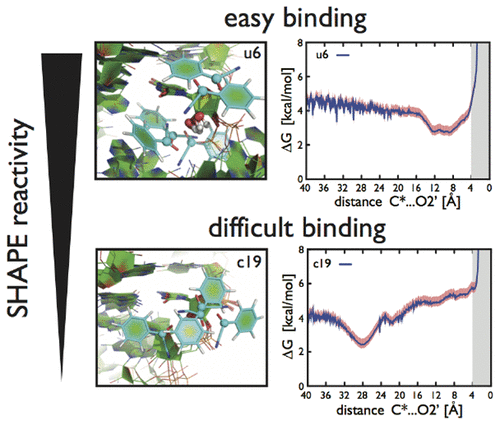
The function of RNA molecules usually depends on their overall fold and on the presence of specific structural motifs. Chemical probing methods are routinely used in combination with nearest-neighbor models to determine RNA secondary structure. Among the available methods, SHAPE is relevant due to its capability to probe all RNA nucleotides and the possibility to be used in vivo. However, the structural determinants for SHAPE reactivity and its mechanism of reaction are still unclear. Here molecular dynamics simulations and enhanced sampling techniques are used to predict the accessibility of nucleotide analogs and larger RNA structural motifs to SHAPE reagents. We show that local RNA reconformations are crucial in allowing reagents to reach the 2'-OH group of a particular nucleotide and that sugar pucker is a major structural factor influencing SHAPE reactivity.
中文翻译:

分子动力学模拟揭示了SHAPE试剂结合与RNA灵活性之间的相互作用。
RNA分子的功能通常取决于它们的整体折叠以及特定结构基序的存在。通常将化学探测方法与最近邻模型结合使用以确定RNA二级结构。在可用的方法中,SHAPE具有相关性,因为它具有探测所有RNA核苷酸的能力以及在体内使用的可能性。但是,尚不清楚SHAPE反应性的结构决定因素及其反应机理。在这里,分子动力学模拟和增强的采样技术可用于预测核苷酸类似物和较大的RNA结构基序对SHAPE试剂的可及性。我们表明,局部RNA重组在允许试剂到达2'端至关重要。
更新日期:2018-01-04
中文翻译:

分子动力学模拟揭示了SHAPE试剂结合与RNA灵活性之间的相互作用。
RNA分子的功能通常取决于它们的整体折叠以及特定结构基序的存在。通常将化学探测方法与最近邻模型结合使用以确定RNA二级结构。在可用的方法中,SHAPE具有相关性,因为它具有探测所有RNA核苷酸的能力以及在体内使用的可能性。但是,尚不清楚SHAPE反应性的结构决定因素及其反应机理。在这里,分子动力学模拟和增强的采样技术可用于预测核苷酸类似物和较大的RNA结构基序对SHAPE试剂的可及性。我们表明,局部RNA重组在允许试剂到达2'端至关重要。



























 京公网安备 11010802027423号
京公网安备 11010802027423号