当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Genetic Toolbox for Modulating the Expression of Heterologous Genes in the Cyanobacterium Synechocystis sp. PCC 6803
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2017-12-22 00:00:00 , DOI: 10.1021/acssynbio.7b00297 Bo Wang 1 , Carrie Eckert 1, 2 , Pin-Ching Maness 1 , Jianping Yu 1
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2017-12-22 00:00:00 , DOI: 10.1021/acssynbio.7b00297 Bo Wang 1 , Carrie Eckert 1, 2 , Pin-Ching Maness 1 , Jianping Yu 1
Affiliation
|
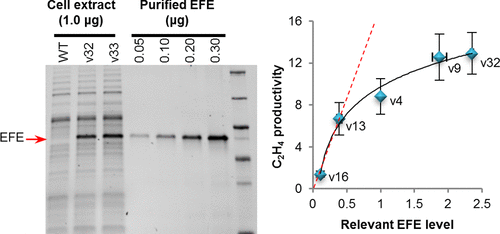
Cyanobacteria, genetic models for photosynthesis research for decades, have recently become attractive hosts for producing renewable fuels and chemicals, owing to their genetic tractability, relatively fast growth, and their ability to utilize sunlight, fix carbon dioxide, and in some cases, fix nitrogen. Despite significant advances, there is still an urgent demand for synthetic biology tools in order to effectively manipulate genetic circuits in cyanobacteria. In this study, we have compared a total of 17 natural and chimeric promoters, focusing on expression of the ethylene-forming enzyme (EFE) in the cyanobacterium Synechocystis sp. PCC 6803. We report the finding that the E. coli σ70 promoter Ptrc is superior compared to the previously reported strong promoters, such as PcpcB and PpsbA, for the expression of EFE. In addition, we found that the EFE expression level was very sensitive to the 5′-untranslated region upstream of the open reading frame. A library of ribosome binding sites (RBSs) was rationally designed and was built and systematically characterized. We demonstrate a strategy complementary to the RBS prediction software to facilitate the rational design of an RBS library to optimize the gene expression in cyanobacteria. Our results show that the EFE expression level is dramatically enhanced through these synthetic biology tools and is no longer the rate-limiting step for cyanobacterial ethylene production. These systematically characterized promoters and the RBS design strategy can serve as useful tools to tune gene expression levels and to identify and mitigate metabolic bottlenecks in cyanobacteria.
中文翻译:

遗传工具箱调制异源基因在蓝藻中的表达蓝藻藻。PCC 6803
蓝细菌是数十年来进行光合作用研究的遗传模型,由于其遗传易处理性,相对较快的生长以及利用阳光,固定二氧化碳以及在某些情况下固定氮的能力,蓝细菌最近已成为生产可再生燃料和化学物质的有吸引力的宿主。 。尽管取得了重大进展,但仍然迫切需要合成生物学工具,以有效地操纵蓝细菌中的遗传回路。在这项研究中,我们已经比较了总共17个天然和嵌合启动子,重点是在蓝藻集胞藻(Synechocystis sp。)中乙烯形成酶(EFE)的表达。PCC 6803我们报告的发现,即大肠杆菌σ 70与以前报道的强启动子(例如PcpcB和PpsbA)相比,Ptrc启动子在EFE的表达上更胜一筹。此外,我们发现EFE表达水平对开放阅读框上游5'-非翻译区非常敏感。核糖体结合位点(RBS)的库是经过合理设计,构建和系统表征的。我们展示了一种补充RBS预测软件的策略,以促进RBS库的合理设计,以优化蓝细菌中的基因表达。我们的结果表明,通过这些合成生物学工具,EFE的表达水平得到了显着提高,并且不再是蓝藻乙烯生产的限速步骤。
更新日期:2017-12-22
中文翻译:

遗传工具箱调制异源基因在蓝藻中的表达蓝藻藻。PCC 6803
蓝细菌是数十年来进行光合作用研究的遗传模型,由于其遗传易处理性,相对较快的生长以及利用阳光,固定二氧化碳以及在某些情况下固定氮的能力,蓝细菌最近已成为生产可再生燃料和化学物质的有吸引力的宿主。 。尽管取得了重大进展,但仍然迫切需要合成生物学工具,以有效地操纵蓝细菌中的遗传回路。在这项研究中,我们已经比较了总共17个天然和嵌合启动子,重点是在蓝藻集胞藻(Synechocystis sp。)中乙烯形成酶(EFE)的表达。PCC 6803我们报告的发现,即大肠杆菌σ 70与以前报道的强启动子(例如PcpcB和PpsbA)相比,Ptrc启动子在EFE的表达上更胜一筹。此外,我们发现EFE表达水平对开放阅读框上游5'-非翻译区非常敏感。核糖体结合位点(RBS)的库是经过合理设计,构建和系统表征的。我们展示了一种补充RBS预测软件的策略,以促进RBS库的合理设计,以优化蓝细菌中的基因表达。我们的结果表明,通过这些合成生物学工具,EFE的表达水平得到了显着提高,并且不再是蓝藻乙烯生产的限速步骤。



























 京公网安备 11010802027423号
京公网安备 11010802027423号