当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Data-Driven Perspective on the Hierarchical Assembly of Molecular Structures
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2017-12-22 00:00:00 , DOI: 10.1021/acs.jctc.7b00990 Lorenzo Boninsegna 1 , Ralf Banisch 2 , Cecilia Clementi 1, 2
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2017-12-22 00:00:00 , DOI: 10.1021/acs.jctc.7b00990 Lorenzo Boninsegna 1 , Ralf Banisch 2 , Cecilia Clementi 1, 2
Affiliation
|
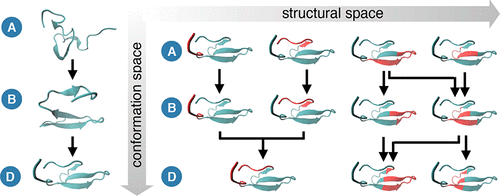
Macromolecular systems are composed of a very large number of atomic degrees of freedom. There is strong evidence suggesting that structural changes occurring in large biomolecular systems at long time scale dynamics may be captured by models coarser than atomistic, although a suitable or optimal coarse-graining is a priori unknown. Here we propose a systematic approach to learning a coarse representation of a macromolecule from microscopic simulation data. In particular, the definition of effective coarse variables is achieved by partitioning the degrees of freedom both in the structural (physical) space and in the conformational space. The identification of groups of microscopic particles forming dynamical coherent states in different metastable states leads to a multiscale description of the system, in space and time. The application of this approach to the folding dynamics of two proteins provides a revised view of the classical idea of prestructured regions (foldons) that combine during a protein-folding process and suggests a hierarchical characterization of the assembly process of folded structures.
中文翻译:

分子结构分层组装的数据驱动视角
大分子系统由非常多的原子自由度组成。有强有力的证据表明,尽管比以往任何时候都合适或最优化的粗粒度模型,但比原子模型更粗糙的模型可能会捕获大型生物分子系统在长时间尺度上发生的结构变化。在这里,我们提出了一种从微观模拟数据中学习大分子粗略表示的系统方法。特别地,通过在结构(物理)空间和构象空间中划分自由度来获得有效的粗略变量的定义。在不同的亚稳态下形成动态相干态的微观粒子群的识别导致了该系统在空间和时间上的多尺度描述。
更新日期:2017-12-22
中文翻译:

分子结构分层组装的数据驱动视角
大分子系统由非常多的原子自由度组成。有强有力的证据表明,尽管比以往任何时候都合适或最优化的粗粒度模型,但比原子模型更粗糙的模型可能会捕获大型生物分子系统在长时间尺度上发生的结构变化。在这里,我们提出了一种从微观模拟数据中学习大分子粗略表示的系统方法。特别地,通过在结构(物理)空间和构象空间中划分自由度来获得有效的粗略变量的定义。在不同的亚稳态下形成动态相干态的微观粒子群的识别导致了该系统在空间和时间上的多尺度描述。


























 京公网安备 11010802027423号
京公网安备 11010802027423号