当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Determinants and Prediction of Esterase Substrate Promiscuity Patterns
ACS Chemical Biology ( IF 4 ) Pub Date : 2017-11-28 00:00:00 , DOI: 10.1021/acschembio.7b00996 Mónica Martínez-Martínez 1 , Cristina Coscolín 1 , Gerard Santiago 2 , Jennifer Chow 3 , Peter J. Stogios 4 , Rafael Bargiela 1 , Christoph Gertler 5 , José Navarro-Fernández 1 , Alexander Bollinger 6 , Stephan Thies 6 , Celia Méndez-García 7 , Ana Popovic 4 , Greg Brown 4 , Tatyana N. Chernikova 5 , Antonio García-Moyano 8 , Gro E. K. Bjerga 8 , Pablo Pérez-García 3 , Tran Hai 5 , Mercedes V. Del Pozo 1 , Runar Stokke 9 , Ida H. Steen 9 , Hong Cui 4 , Xiaohui Xu 4 , Boguslaw P. Nocek 10 , María Alcaide 1 , Marco Distaso 5 , Victoria Mesa 7 , Ana I. Peláez 7 , Jesús Sánchez 7 , Patrick C. F. Buchholz 11 , Jürgen Pleiss 11 , Antonio Fernández-Guerra 12, 13, 14 , Frank O. Glöckner 12, 13 , Olga V. Golyshina 5 , Michail M. Yakimov 15, 16 , Alexei Savchenko 4 , Karl-Erich Jaeger 6, 17 , Alexander F. Yakunin 4 , Wolfgang R. Streit 3 , Peter N. Golyshin 5 , Víctor Guallar 2, 18 , Manuel Ferrer 1 , The INMARE Consortium
ACS Chemical Biology ( IF 4 ) Pub Date : 2017-11-28 00:00:00 , DOI: 10.1021/acschembio.7b00996 Mónica Martínez-Martínez 1 , Cristina Coscolín 1 , Gerard Santiago 2 , Jennifer Chow 3 , Peter J. Stogios 4 , Rafael Bargiela 1 , Christoph Gertler 5 , José Navarro-Fernández 1 , Alexander Bollinger 6 , Stephan Thies 6 , Celia Méndez-García 7 , Ana Popovic 4 , Greg Brown 4 , Tatyana N. Chernikova 5 , Antonio García-Moyano 8 , Gro E. K. Bjerga 8 , Pablo Pérez-García 3 , Tran Hai 5 , Mercedes V. Del Pozo 1 , Runar Stokke 9 , Ida H. Steen 9 , Hong Cui 4 , Xiaohui Xu 4 , Boguslaw P. Nocek 10 , María Alcaide 1 , Marco Distaso 5 , Victoria Mesa 7 , Ana I. Peláez 7 , Jesús Sánchez 7 , Patrick C. F. Buchholz 11 , Jürgen Pleiss 11 , Antonio Fernández-Guerra 12, 13, 14 , Frank O. Glöckner 12, 13 , Olga V. Golyshina 5 , Michail M. Yakimov 15, 16 , Alexei Savchenko 4 , Karl-Erich Jaeger 6, 17 , Alexander F. Yakunin 4 , Wolfgang R. Streit 3 , Peter N. Golyshin 5 , Víctor Guallar 2, 18 , Manuel Ferrer 1 , The INMARE Consortium
Affiliation
|
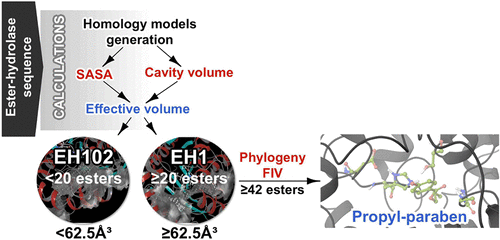
Esterases receive special attention because of their wide distribution in biological systems and environments and their importance for physiology and chemical synthesis. The prediction of esterases’ substrate promiscuity level from sequence data and the molecular reasons why certain such enzymes are more promiscuous than others remain to be elucidated. This limits the surveillance of the sequence space for esterases potentially leading to new versatile biocatalysts and new insights into their role in cellular function. Here, we performed an extensive analysis of the substrate spectra of 145 phylogenetically and environmentally diverse microbial esterases, when tested with 96 diverse esters. We determined the primary factors shaping their substrate range by analyzing substrate range patterns in combination with structural analysis and protein–ligand simulations. We found a structural parameter that helps rank (classify) the promiscuity level of esterases from sequence data at 94% accuracy. This parameter, the active site effective volume, exemplifies the topology of the catalytic environment by measuring the active site cavity volume corrected by the relative solvent accessible surface area (SASA) of the catalytic triad. Sequences encoding esterases with active site effective volumes (cavity volume/SASA) above a threshold show greater substrate spectra, which can be further extended in combination with phylogenetic data. This measure provides also a valuable tool for interrogating substrates capable of being converted. This measure, found to be transferred to phosphatases of the haloalkanoic acid dehalogenase superfamily and possibly other enzymatic systems, represents a powerful tool for low-cost bioprospecting for esterases with broad substrate ranges, in large scale sequence data sets.
中文翻译:

酯酶底物混杂模式的决定因素和预测
酯酶由于其在生物系统和环境中的广泛分布及其对生理学和化学合成的重要性而受到特别关注。从序列数据预测酯酶底物混杂水平的原因以及某些这种酶比其他酶混杂的分子原因还有待阐明。这限制了对酯酶序列空间的监视,从而潜在地导致了新的多功能生物催化剂及其对它们在细胞功能中作用的新见解。在这里,当用96种不同的酯类进行测试时,我们对145种系统发育和环境多样的微生物酯酶的底物谱进行了广泛的分析。通过结合结构分析和蛋白质-配体模拟分析底物范围模式,我们确定了影响其底物范围的主要因素。我们发现了一个结构参数,可帮助从序列数据中以94%的准确性对酯酶的混杂程度进行排名(分类)。该参数是活性位有效体积,通过测量由催化三元组的相对溶剂可及表面积(SASA)校正的活性位腔体积来举例说明催化环境的拓扑。具有高于阈值的活性位点有效体积(空洞体积/ SASA)的编码酯酶的序列显示出更大的底物光谱,可以结合系统发育数据进一步扩展该底物光谱。该措施还提供了一种有价值的工具,用于询问能够被转换的衬底。这项措施
更新日期:2017-11-28
中文翻译:

酯酶底物混杂模式的决定因素和预测
酯酶由于其在生物系统和环境中的广泛分布及其对生理学和化学合成的重要性而受到特别关注。从序列数据预测酯酶底物混杂水平的原因以及某些这种酶比其他酶混杂的分子原因还有待阐明。这限制了对酯酶序列空间的监视,从而潜在地导致了新的多功能生物催化剂及其对它们在细胞功能中作用的新见解。在这里,当用96种不同的酯类进行测试时,我们对145种系统发育和环境多样的微生物酯酶的底物谱进行了广泛的分析。通过结合结构分析和蛋白质-配体模拟分析底物范围模式,我们确定了影响其底物范围的主要因素。我们发现了一个结构参数,可帮助从序列数据中以94%的准确性对酯酶的混杂程度进行排名(分类)。该参数是活性位有效体积,通过测量由催化三元组的相对溶剂可及表面积(SASA)校正的活性位腔体积来举例说明催化环境的拓扑。具有高于阈值的活性位点有效体积(空洞体积/ SASA)的编码酯酶的序列显示出更大的底物光谱,可以结合系统发育数据进一步扩展该底物光谱。该措施还提供了一种有价值的工具,用于询问能够被转换的衬底。这项措施



























 京公网安备 11010802027423号
京公网安备 11010802027423号