当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
pSite: Amino Acid Confidence Evaluation for Quality Control of De Novo Peptide Sequencing and Modification Site Localization
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acs.jproteome.7b00428 Hao Yang 1, 2 , Hao Chi 1 , Wen-Jing Zhou 1, 2 , Wen-Feng Zeng 1, 2 , Chao Liu 1 , Rui-Min Wang 1, 2 , Zhao-Wei Wang 1, 2 , Xiu-Nan Niu 1, 2 , Zhen-Lin Chen 1, 2 , Si-Min He 1, 2
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acs.jproteome.7b00428 Hao Yang 1, 2 , Hao Chi 1 , Wen-Jing Zhou 1, 2 , Wen-Feng Zeng 1, 2 , Chao Liu 1 , Rui-Min Wang 1, 2 , Zhao-Wei Wang 1, 2 , Xiu-Nan Niu 1, 2 , Zhen-Lin Chen 1, 2 , Si-Min He 1, 2
Affiliation
|
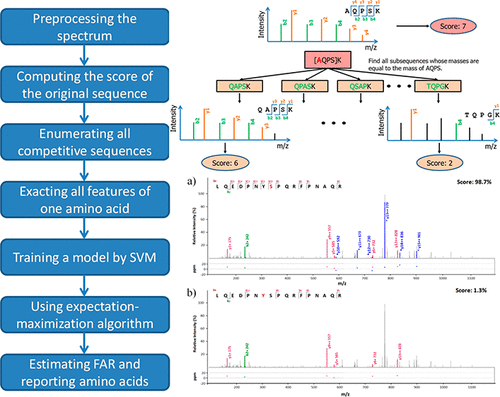
MS-based de novo peptide sequencing has been improved remarkably with significant development of mass-spectrometry and computational approaches but still lacks quality-control methods. Here we proposed a novel algorithm pSite to evaluate the confidence of each amino acid rather than the full-length peptides obtained by de novo peptide sequencing. A semi-supervised learning approach was used to discriminate correct amino acids from random one; then, an expectation-maximization algorithm was used to adaptively control the false amino-acid rate (FAR). On three test data sets, pSite recalled 86% more amino acids on average than PEAKS at the FAR of 5%.
中文翻译:

pSite:用于从头肽测序和修饰位点定位的质量控制的氨基酸置信度评估
随着质谱和计算方法的重大发展,基于质谱的从头肽测序已得到显着改善,但仍缺乏质量控制方法。在这里,我们提出了一种新颖的算法pSite来评估每个氨基酸的可信度,而不是通过从头测序获得的全长肽的可信度。使用半监督学习方法将正确的氨基酸与随机的氨基酸区分开。然后,采用期望最大化算法自适应地控制假氨基酸率(FAR)。在三个测试数据集上,pSite在5%的FAR时召回的氨基酸平均比PEAKS多86%。
更新日期:2017-11-22
中文翻译:

pSite:用于从头肽测序和修饰位点定位的质量控制的氨基酸置信度评估
随着质谱和计算方法的重大发展,基于质谱的从头肽测序已得到显着改善,但仍缺乏质量控制方法。在这里,我们提出了一种新颖的算法pSite来评估每个氨基酸的可信度,而不是通过从头测序获得的全长肽的可信度。使用半监督学习方法将正确的氨基酸与随机的氨基酸区分开。然后,采用期望最大化算法自适应地控制假氨基酸率(FAR)。在三个测试数据集上,pSite在5%的FAR时召回的氨基酸平均比PEAKS多86%。


























 京公网安备 11010802027423号
京公网安备 11010802027423号