当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Proteomic Characterization of Transcription and Splicing Factors Associated with a Metastatic Phenotype in Colorectal Cancer
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acs.jproteome.7b00548 Sofía Torres 1 , Irene García-Palmero 1 , Consuelo Marín-Vicente 1, 2 , Rubén A. Bartolomé 1 , Eva Calviño 1 , María Jesús Fernández-Aceñero 3 , J. Ignacio Casal 1
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acs.jproteome.7b00548 Sofía Torres 1 , Irene García-Palmero 1 , Consuelo Marín-Vicente 1, 2 , Rubén A. Bartolomé 1 , Eva Calviño 1 , María Jesús Fernández-Aceñero 3 , J. Ignacio Casal 1
Affiliation
|
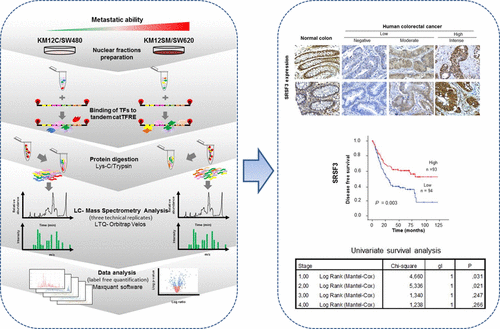
We investigated new transcription and splicing factors associated with the metastatic phenotype in colorectal cancer. A concatenated tandem array of consensus transcription factor (TF)-response elements was used to pull down nuclear extracts in two different pairs of colorectal cancer cells, KM12SM/KM12C and SW620/480, genetically related but differing in metastatic ability. Proteins were analyzed by label-free LC–MS and quantified with MaxLFQ. We found 240 proteins showing a significant dysregulation in highly metastatic KM12SM cells relative to nonmetastatic KM12C cells and 257 proteins in metastatic SW620 versus SW480. In both cell lines there were similar alterations in genuine TFs and components of the splicing machinery like UPF1, TCF7L2/TCF-4, YBX1, or SRSF3. However, a significant number of alterations were cell-line specific. Functional silencing of MAFG, TFE3, TCF7L2/TCF-4, and SRSF3 in KM12 cells caused alterations in adhesion, survival, proliferation, migration, and liver homing, supporting their role in metastasis. Finally, we investigated the prognostic value of the altered TFs and splicing factors in cancer patients. SRSF3 and SFPQ showed significant prognostic value. We observed that SRSF3 displayed a gradual loss of expression associated with cancer progression. Loss of SRSF3 expression was significantly associated with poor survival and shorter disease-free survival, particularly in early stages, in colorectal cancer.
中文翻译:

蛋白质组学表征的转录和剪接因子与大肠癌转移表型有关。
我们调查了与大肠癌转移表型相关的新转录和剪接因子。串联连接的共有转录因子(TF)反应元件串联阵列可拉低两对大肠癌细胞KM12SM / KM12C和SW620 / 480的遗传相关但转移能力不同的核提取物。通过无标记LC-MS分析蛋白质,并用MaxLFQ进行定量。我们发现,相对于非转移性KM12C细胞,在高度转移的KM12SM细胞中有240种蛋白显示出明显的失调,而在转移SW620与SW480中,则有257种蛋白。在两种细胞系中,真正的TF和剪接机器的组件(如UPF1,TCF7L2 / TCF-4,YBX1或SRSF3)都有类似的变化。然而,许多改变是细胞系特异性的。KM12细胞中MAFG,TFE3,TCF7L2 / TCF-4和SRSF3的功能沉默导致粘附,存活,增殖,迁移和肝归巢改变,从而支持其在转移中的作用。最后,我们调查了改变的TFs和剪接因子在癌症患者中的预后价值。SRSF3和SFPQ显示出显着的预后价值。我们观察到,SRSF3显示出与癌症进展相关的表达逐渐丧失。SRSF3表达的丧失与大肠癌的不良生存率和较短的无病生存率显着相关,尤其是在早期阶段。我们调查了改变的TFs和剪接因子在癌症患者中的预后价值。SRSF3和SFPQ显示出显着的预后价值。我们观察到,SRSF3显示出与癌症进展相关的表达逐渐丧失。SRSF3表达的丧失与大肠癌的不良生存率和较短的无病生存率显着相关,尤其是在早期阶段。我们调查了改变的TFs和剪接因子在癌症患者中的预后价值。SRSF3和SFPQ显示出显着的预后价值。我们观察到,SRSF3显示出与癌症进展相关的表达逐渐丧失。SRSF3表达的丧失与大肠癌的不良生存率和较短的无病生存率显着相关,尤其是在早期阶段。
更新日期:2017-11-22
中文翻译:

蛋白质组学表征的转录和剪接因子与大肠癌转移表型有关。
我们调查了与大肠癌转移表型相关的新转录和剪接因子。串联连接的共有转录因子(TF)反应元件串联阵列可拉低两对大肠癌细胞KM12SM / KM12C和SW620 / 480的遗传相关但转移能力不同的核提取物。通过无标记LC-MS分析蛋白质,并用MaxLFQ进行定量。我们发现,相对于非转移性KM12C细胞,在高度转移的KM12SM细胞中有240种蛋白显示出明显的失调,而在转移SW620与SW480中,则有257种蛋白。在两种细胞系中,真正的TF和剪接机器的组件(如UPF1,TCF7L2 / TCF-4,YBX1或SRSF3)都有类似的变化。然而,许多改变是细胞系特异性的。KM12细胞中MAFG,TFE3,TCF7L2 / TCF-4和SRSF3的功能沉默导致粘附,存活,增殖,迁移和肝归巢改变,从而支持其在转移中的作用。最后,我们调查了改变的TFs和剪接因子在癌症患者中的预后价值。SRSF3和SFPQ显示出显着的预后价值。我们观察到,SRSF3显示出与癌症进展相关的表达逐渐丧失。SRSF3表达的丧失与大肠癌的不良生存率和较短的无病生存率显着相关,尤其是在早期阶段。我们调查了改变的TFs和剪接因子在癌症患者中的预后价值。SRSF3和SFPQ显示出显着的预后价值。我们观察到,SRSF3显示出与癌症进展相关的表达逐渐丧失。SRSF3表达的丧失与大肠癌的不良生存率和较短的无病生存率显着相关,尤其是在早期阶段。我们调查了改变的TFs和剪接因子在癌症患者中的预后价值。SRSF3和SFPQ显示出显着的预后价值。我们观察到,SRSF3显示出与癌症进展相关的表达逐渐丧失。SRSF3表达的丧失与大肠癌的不良生存率和较短的无病生存率显着相关,尤其是在早期阶段。


























 京公网安备 11010802027423号
京公网安备 11010802027423号