当前位置:
X-MOL 学术
›
ACS Cent. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Prediction of New Stabilizing Mutations Based on Mechanistic Insights from Markov State Models
ACS Central Science ( IF 18.2 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acscentsci.7b00465 Maxwell I. Zimmerman 1 , Kathryn M. Hart 1 , Carrie A. Sibbald 1 , Thomas E. Frederick 1 , John R. Jimah 2 , Catherine R. Knoverek 1 , Niraj H. Tolia 1, 2 , Gregory R. Bowman 1, 3
ACS Central Science ( IF 18.2 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acscentsci.7b00465 Maxwell I. Zimmerman 1 , Kathryn M. Hart 1 , Carrie A. Sibbald 1 , Thomas E. Frederick 1 , John R. Jimah 2 , Catherine R. Knoverek 1 , Niraj H. Tolia 1, 2 , Gregory R. Bowman 1, 3
Affiliation
|
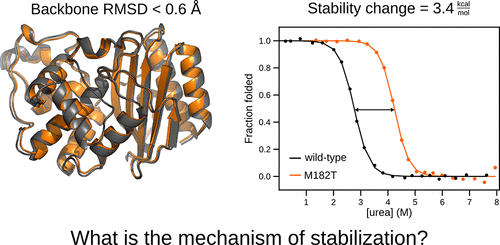
Protein stabilization is fundamental to enzyme function and evolution, yet understanding the determinants of a protein’s stability remains a challenge. This is largely due to a shortage of atomically detailed models for the ensemble of relevant protein conformations and their relative populations. For example, the M182T substitution in TEM β-lactamase, an enzyme that confers antibiotic resistance to bacteria, is stabilizing but the precise mechanism remains unclear. Here, we employ Markov state models (MSMs) to uncover how M182T shifts the distribution of different structures that TEM adopts. We find that M182T stabilizes a helix that is a key component of a domain interface. We then predict the effects of other mutations, including a novel stabilizing mutation, and experimentally test our predictions using a combination of stability measurements, crystallography, NMR, and in vivo measurements of bacterial fitness. We expect our insights and methodology to provide a valuable foundation for protein design.
中文翻译:

基于马尔可夫状态模型的机械洞察力预测新的稳定突变
蛋白质稳定化是酶功能和进化的基础,但是了解蛋白质稳定性的决定因素仍然是一个挑战。这主要是由于缺乏相关蛋白质构象及其相对种群的原子详细模型的缺乏。例如,TEMβ-内酰胺酶中的M182T取代是一种稳定抗生素的酶,该酶可赋予细菌对细菌的抗药性,但确切的机制尚不清楚。在这里,我们采用马尔可夫状态模型(MSM)来揭示M182T如何改变TEM采用的不同结构的分布。我们发现M182T稳定了作为域接口的关键组件的螺旋。然后,我们预测其他突变(包括新的稳定突变)的影响,并通过结合使用稳定性测量来实验性地测试我们的预测,体内细菌适应性测量。我们希望我们的见解和方法学能够为蛋白质设计提供有价值的基础。
更新日期:2017-11-21
中文翻译:

基于马尔可夫状态模型的机械洞察力预测新的稳定突变
蛋白质稳定化是酶功能和进化的基础,但是了解蛋白质稳定性的决定因素仍然是一个挑战。这主要是由于缺乏相关蛋白质构象及其相对种群的原子详细模型的缺乏。例如,TEMβ-内酰胺酶中的M182T取代是一种稳定抗生素的酶,该酶可赋予细菌对细菌的抗药性,但确切的机制尚不清楚。在这里,我们采用马尔可夫状态模型(MSM)来揭示M182T如何改变TEM采用的不同结构的分布。我们发现M182T稳定了作为域接口的关键组件的螺旋。然后,我们预测其他突变(包括新的稳定突变)的影响,并通过结合使用稳定性测量来实验性地测试我们的预测,体内细菌适应性测量。我们希望我们的见解和方法学能够为蛋白质设计提供有价值的基础。



























 京公网安备 11010802027423号
京公网安备 11010802027423号