当前位置:
X-MOL 学术
›
Mol. Biosyst.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Computational identification of protein S-sulfenylation sites by incorporating the multiple sequence features information
Molecular BioSystems Pub Date : 2017-09-18 00:00:00 , DOI: 10.1039/c7mb00491e Md. Mehedi Hasan 1, 2, 3, 4 , Dianjing Guo 5, 6, 7, 8 , Hiroyuki Kurata 1, 2, 3, 4, 9
Molecular BioSystems Pub Date : 2017-09-18 00:00:00 , DOI: 10.1039/c7mb00491e Md. Mehedi Hasan 1, 2, 3, 4 , Dianjing Guo 5, 6, 7, 8 , Hiroyuki Kurata 1, 2, 3, 4, 9
Affiliation
|
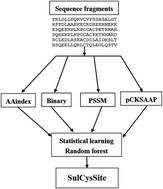
Cysteine S-sulfenylation is a major type of posttranslational modification that contributes to protein structure and function regulation in many cellular processes. Experimental identification of S-sulfenylation sites is challenging, due to the low abundance of proteins and the inefficient experimental methods. Computational identification of S-sulfenylation sites is an alternative strategy to annotate the S-sulfenylated proteome. In this study, a novel computational predictor SulCysSite was developed for accurate prediction of S-sulfenylation sites based on multiple sequence features, including amino acid index properties, binary amino acid codes, position specific scoring matrix, and compositions of profile-based amino acids. To learn the prediction model of SulCysSite, a random forest classifier was applied. The final SulCysSite achieved an AUC value of 0.819 in a 10-fold cross-validation test. It also exhibited higher performance than other existing computational predictors. In addition, the hidden and complex mechanisms were extracted from the predictive model of SulCysSite to investigate the understandable rules (i.e. feature combination) of S-sulfenylation sites. The SulCysSite is a useful computational resource for prediction of S-sulfenylation sites. The online interface and datasets are publicly available at http://kurata14.bio.kyutech.ac.jp/SulCysSite/.
中文翻译:

通过结合多个序列特征信息的蛋白质S-亚磺酰化位点的计算鉴定
半胱氨酸S-亚磺酰基化是翻译后修饰的主要类型,其在许多细胞过程中有助于蛋白质结构和功能调节。由于蛋白质丰度低和实验方法效率低下,S-亚磺酰基化位点的实验鉴定具有挑战性。S-亚磺酰基化位点的计算鉴定是注释S-亚磺酰基化蛋白质组的另一种策略。在这项研究中,开发了一种新颖的计算预测因子SulCysSite,用于基于多个序列特征(包括氨基酸索引特性,二元氨基酸代码,位置特异性得分矩阵和基于轮廓的氨基酸的组成)来准确预测S-亚磺酰基化位点。为了学习SulCysSite的预测模型,使用了随机森林分类器。最终的SulCysSite在10倍交叉验证测试中达到了0.819的AUC值。它也表现出比其他现有的计算预测器更高的性能。此外,还从SulCysSite的预测模型中提取了隐藏的复杂机制,以研究可理解的规则(即特征组合)的S-亚磺酰基化位点。SulCysSite是有用的计算资源,可用于预测S-亚磺酰化位点。在线界面和数据集可在http://kurata14.bio.kyutech.ac.jp/SulCysSite/上公开获得。
更新日期:2017-11-21
中文翻译:

通过结合多个序列特征信息的蛋白质S-亚磺酰化位点的计算鉴定
半胱氨酸S-亚磺酰基化是翻译后修饰的主要类型,其在许多细胞过程中有助于蛋白质结构和功能调节。由于蛋白质丰度低和实验方法效率低下,S-亚磺酰基化位点的实验鉴定具有挑战性。S-亚磺酰基化位点的计算鉴定是注释S-亚磺酰基化蛋白质组的另一种策略。在这项研究中,开发了一种新颖的计算预测因子SulCysSite,用于基于多个序列特征(包括氨基酸索引特性,二元氨基酸代码,位置特异性得分矩阵和基于轮廓的氨基酸的组成)来准确预测S-亚磺酰基化位点。为了学习SulCysSite的预测模型,使用了随机森林分类器。最终的SulCysSite在10倍交叉验证测试中达到了0.819的AUC值。它也表现出比其他现有的计算预测器更高的性能。此外,还从SulCysSite的预测模型中提取了隐藏的复杂机制,以研究可理解的规则(即特征组合)的S-亚磺酰基化位点。SulCysSite是有用的计算资源,可用于预测S-亚磺酰化位点。在线界面和数据集可在http://kurata14.bio.kyutech.ac.jp/SulCysSite/上公开获得。


























 京公网安备 11010802027423号
京公网安备 11010802027423号