当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
An Automated Image Analysis Method for Segmenting Fluorescent Bacteria in Three Dimensions
Biochemistry ( IF 2.9 ) Pub Date : 2017-11-10 00:00:00 , DOI: 10.1021/acs.biochem.7b00839 Matthew A. Reyer 1 , Eric L. McLean 1 , Shriram Chennakesavalu 1 , Jingyi Fei 1
Biochemistry ( IF 2.9 ) Pub Date : 2017-11-10 00:00:00 , DOI: 10.1021/acs.biochem.7b00839 Matthew A. Reyer 1 , Eric L. McLean 1 , Shriram Chennakesavalu 1 , Jingyi Fei 1
Affiliation
|
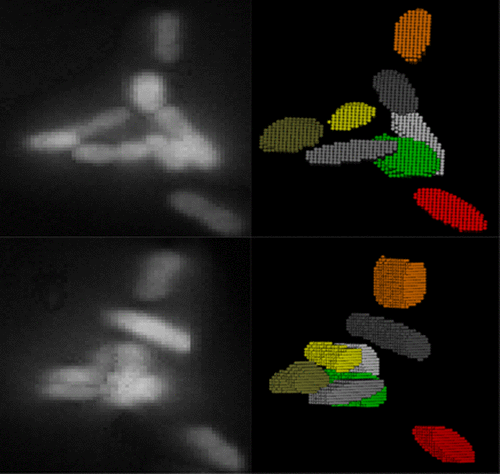
Single-cell fluorescence imaging is a powerful technique for studying inherently heterogeneous biological processes. To correlate a genotype or phenotype to a specific cell, images containing a population of cells must first be properly segmented. However, a proper segmentation with minimal user input becomes challenging when cells are clustered or overlapping in three dimensions. We introduce a new analysis package, Seg-3D, for the segmentation of bacterial cells in three-dimensional (3D) images, based on local thresholding, shape analysis, concavity-based cluster splitting, and morphology-based 3D reconstruction. The reconstructed cell volumes allow us to directly quantify the fluorescent signals from biomolecules of interest within individual cells. We demonstrate the application of this analysis package in 3D segmentation of individual bacterial pathogens invading host cells. We believe Seg-3D can be an efficient and simple program that can be used to analyze a wide variety of single-cell images, especially for biological systems involving random 3D orientation and clustering behavior, such as bacterial infection or colonization.
中文翻译:

一种三维三维荧光细菌自动分割方法
单细胞荧光成像是研究固有的异质生物学过程的强大技术。要将基因型或表型与特定细胞相关联,必须首先正确分割包含细胞群的图像。但是,当单元在三个维度上成簇或重叠时,用最少的用户输入进行正确的分割就变得很困难。我们基于局部阈值,形状分析,基于凹度的聚类分裂和基于形态学的3D重建,引入了一种新的分析程序包Seg-3D,用于在三维(3D)图像中分割细菌细胞。重建的细胞体积使我们能够直接量化单个细胞内目标生物分子的荧光信号。我们证明了此分析包在入侵宿主细胞的单个细菌病原体的3D分割中的应用。我们相信Seg-3D可以是一种高效且简单的程序,可用于分析各种单细胞图像,尤其是对于涉及随机3D方向和聚集行为(例如细菌感染或定植)的生物系统。
更新日期:2017-11-11
中文翻译:

一种三维三维荧光细菌自动分割方法
单细胞荧光成像是研究固有的异质生物学过程的强大技术。要将基因型或表型与特定细胞相关联,必须首先正确分割包含细胞群的图像。但是,当单元在三个维度上成簇或重叠时,用最少的用户输入进行正确的分割就变得很困难。我们基于局部阈值,形状分析,基于凹度的聚类分裂和基于形态学的3D重建,引入了一种新的分析程序包Seg-3D,用于在三维(3D)图像中分割细菌细胞。重建的细胞体积使我们能够直接量化单个细胞内目标生物分子的荧光信号。我们证明了此分析包在入侵宿主细胞的单个细菌病原体的3D分割中的应用。我们相信Seg-3D可以是一种高效且简单的程序,可用于分析各种单细胞图像,尤其是对于涉及随机3D方向和聚集行为(例如细菌感染或定植)的生物系统。



























 京公网安备 11010802027423号
京公网安备 11010802027423号