当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Large Scale Mutational and Kinetic Analysis of a Self-Hydrolyzing Deoxyribozyme
ACS Chemical Biology ( IF 4 ) Pub Date : 2017-11-03 00:00:00 , DOI: 10.1021/acschembio.7b00621 V. Dhamodharan 1 , Shungo Kobori 1 , Yohei Yokobayashi 1
ACS Chemical Biology ( IF 4 ) Pub Date : 2017-11-03 00:00:00 , DOI: 10.1021/acschembio.7b00621 V. Dhamodharan 1 , Shungo Kobori 1 , Yohei Yokobayashi 1
Affiliation
|
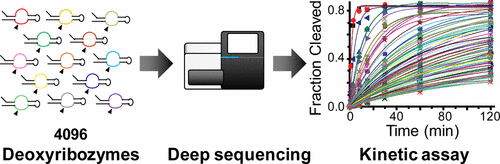
Deoxyribozymes are catalytic DNA sequences whose atomic structures are generally difficult to elucidate. Mutational analysis remains a principal approach for understanding and engineering deoxyribozymes with diverse catalytic activities. However, laborious preparation and biochemical characterization of individual sequences severely limit the number of mutants that can be studied biochemically. Here, we applied deep sequencing to directly measure the activities of self-hydrolyzing deoxyribozyme sequences in high throughput. First, all single and double mutants within the 15-base catalytic core of the deoxyribozyme I-R3 were assayed to unambiguously determine the tolerated and untolerated mutations at each position. Subsequently, 4096 deoxyribozyme variants with tolerated base substitutions at seven positions were kinetically assayed in parallel. We identified 533 active mutants whose first-order rate constants and activation energies were determined. The results indicate an isolated and narrow peak in the deoxyribozyme sequence space and provide a quantitative view of the effects of multiple mutations on the deoxyribozyme activity for the first time.
中文翻译:

自水解脱氧核酶的大规模突变和动力学分析
脱氧核酶是催化性的DNA序列,其原子结构通常难以阐明。突变分析仍然是理解和工程化具有多种催化活性的脱氧核酶的主要方法。然而,费力的制备和单个序列的生化表征严重限制了可以进行生化研究的突变体的数量。在这里,我们应用深度测序直接测量高通量中自水解脱氧核酶序列的活性。首先,测定脱氧核糖酶I-R3的15个碱基的催化核心内的所有单突变体和双突变体,以明确确定每个位置上的耐受和非耐受突变。随后,平行地动力学分析了在七个位置具有耐受的碱基取代的4096个脱氧核酶变体。我们鉴定了533个活性突变体,它们的一阶速率常数和活化能被确定。结果表明在脱氧核酶序列空间中一个孤立的狭窄峰,并首次提供了多个突变对脱氧核酶活性影响的定量视图。
更新日期:2017-11-05
中文翻译:

自水解脱氧核酶的大规模突变和动力学分析
脱氧核酶是催化性的DNA序列,其原子结构通常难以阐明。突变分析仍然是理解和工程化具有多种催化活性的脱氧核酶的主要方法。然而,费力的制备和单个序列的生化表征严重限制了可以进行生化研究的突变体的数量。在这里,我们应用深度测序直接测量高通量中自水解脱氧核酶序列的活性。首先,测定脱氧核糖酶I-R3的15个碱基的催化核心内的所有单突变体和双突变体,以明确确定每个位置上的耐受和非耐受突变。随后,平行地动力学分析了在七个位置具有耐受的碱基取代的4096个脱氧核酶变体。我们鉴定了533个活性突变体,它们的一阶速率常数和活化能被确定。结果表明在脱氧核酶序列空间中一个孤立的狭窄峰,并首次提供了多个突变对脱氧核酶活性影响的定量视图。

























 京公网安备 11010802027423号
京公网安备 11010802027423号