当前位置:
X-MOL 学术
›
Cell Stem Cell
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
An endosiRNA-Based Repression Mechanism Counteracts Transposon Activation during Global DNA Demethylation in Embryonic Stem Cells.
Cell Stem Cell ( IF 23.9 ) Pub Date : 2017-Nov-02 , DOI: 10.1016/j.stem.2017.10.004 Rebecca V. Berrens , Simon Andrews , Dominik Spensberger , Fátima Santos , Wendy Dean , Poppy Gould , Jafar Sharif , Nelly Olova , Tamir Chandra , Haruhiko Koseki , Ferdinand von Meyenn , Wolf Reik
Cell Stem Cell ( IF 23.9 ) Pub Date : 2017-Nov-02 , DOI: 10.1016/j.stem.2017.10.004 Rebecca V. Berrens , Simon Andrews , Dominik Spensberger , Fátima Santos , Wendy Dean , Poppy Gould , Jafar Sharif , Nelly Olova , Tamir Chandra , Haruhiko Koseki , Ferdinand von Meyenn , Wolf Reik
|
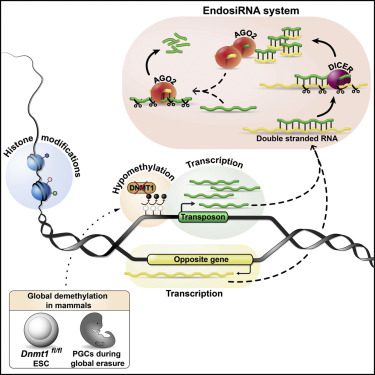
Erasure of DNA methylation and repressive chromatin marks in the mammalian germline leads to risk of transcriptional activation of transposable elements (TEs). Here, we used mouse embryonic stem cells (ESCs) to identify an endosiRNA-based mechanism involved in suppression of TE transcription. In ESCs with DNA demethylation induced by acute deletion of Dnmt1, we saw an increase in sense transcription at TEs, resulting in an abundance of sense/antisense transcripts leading to high levels of ARGONAUTE2 (AGO2)-bound small RNAs. Inhibition of Dicer or Ago2 expression revealed that small RNAs are involved in an immediate response to demethylation-induced transposon activation, while the deposition of repressive histone marks follows as a chronic response. In vivo, we also found TE-specific endosiRNAs present during primordial germ cell development. Our results suggest that antisense TE transcription is a "trap" that elicits an endosiRNA response to restrain acute transposon activity during epigenetic reprogramming in the mammalian germline.
中文翻译:

基于endosiRNA的抑制机制抵消胚胎干细胞中的整体DNA去甲基化过程中的转座子激活。
消除哺乳动物种系中的DNA甲基化和抑制性染色质标记会导致转座因子(TEs)转录激活的风险。在这里,我们使用小鼠胚胎干细胞(ESCs)来识别参与抑制TE转录的基于endsiRNA的机制。在由Dnmt1的急性缺失诱导的DNA去甲基化的ESC,我们看到在TEs的有义转录增加,导致大量的有义/反义转录本,导致高水平的ARGONAUTE2(AGO2)绑定的小RNA。对Dicer或Ago2表达的抑制表明,小分子RNA参与了对去甲基化诱导的转座子激活的即时反应,而抑制性组蛋白标记的沉积则是慢性反应。在体内,我们还发现在原始生殖细胞发育过程中存在TE特异性endosiRNA。
更新日期:2017-11-02
中文翻译:

基于endosiRNA的抑制机制抵消胚胎干细胞中的整体DNA去甲基化过程中的转座子激活。
消除哺乳动物种系中的DNA甲基化和抑制性染色质标记会导致转座因子(TEs)转录激活的风险。在这里,我们使用小鼠胚胎干细胞(ESCs)来识别参与抑制TE转录的基于endsiRNA的机制。在由Dnmt1的急性缺失诱导的DNA去甲基化的ESC,我们看到在TEs的有义转录增加,导致大量的有义/反义转录本,导致高水平的ARGONAUTE2(AGO2)绑定的小RNA。对Dicer或Ago2表达的抑制表明,小分子RNA参与了对去甲基化诱导的转座子激活的即时反应,而抑制性组蛋白标记的沉积则是慢性反应。在体内,我们还发现在原始生殖细胞发育过程中存在TE特异性endosiRNA。



























 京公网安备 11010802027423号
京公网安备 11010802027423号