当前位置:
X-MOL 学术
›
Environ. Sci.: Nano
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Time-dependent bacterial transcriptional response to CuO nanoparticles differs from that of Cu2+ and provides insights into CuO nanoparticle toxicity mechanisms†
Environmental Science: Nano ( IF 7.3 ) Pub Date : 2017-10-20 00:00:00 , DOI: 10.1039/c7en00600d Joe D. Moore 1, 2, 3, 4, 5 , Astrid Avellan 1, 2, 3, 4, 5 , Clinton W. Noack 1, 2, 3, 4 , Yisong Guo 2, 3, 4, 6 , Gregory V. Lowry 1, 2, 3, 4, 5 , Kelvin B. Gregory 1, 2, 3, 4, 5
Environmental Science: Nano ( IF 7.3 ) Pub Date : 2017-10-20 00:00:00 , DOI: 10.1039/c7en00600d Joe D. Moore 1, 2, 3, 4, 5 , Astrid Avellan 1, 2, 3, 4, 5 , Clinton W. Noack 1, 2, 3, 4 , Yisong Guo 2, 3, 4, 6 , Gregory V. Lowry 1, 2, 3, 4, 5 , Kelvin B. Gregory 1, 2, 3, 4, 5
Affiliation
|
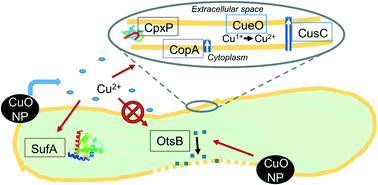
Better understanding of the time-resolved gene expression response of bacteria to CuO engineered nanomaterials (ENM) could lead to an improved mechanistic understanding of their antimicrobial effects. In this study, reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) has been used to characterize a time series of Escherichia coli gene expression after exposure to CuO nanoparticles (NP) (100 mg L−1 as Cu). Two dissolved Cu exposures in concentrations matching observed dissolution from the CuO NPs in the growth medium (1 ppm total Cu after 180 min) were included as a comparison against NP exposure. In the pulse dissolved Cu exposure, 1 ppm Cu was added at the beginning of the experiment. In the gradual dissolved Cu exposure, a total of 1 ppm dissolved Cu was divided into four doses (at 0, 10, 30, and 60 min) that matched the observed CuO NP dissolution. NP exposure led to induction of membrane damage gene expression, which aligns with hyperspectral imaging (HSI) results that identified a high NP affinity for cellular membranes shortly after exposure. Reactive oxygen species (ROS)-responsive genes were not induced for the NP-exposed E. coli within the 60 min time scale where Cu-(copA, cueO, and cusC) and protein damage gene (cpxP) expression were most induced – Cu2+ treatments led to minimal induction. Both Cu2+ treatments (pulse and gradual addition) led to higher levels of Cu- and protein damage-responsive gene induction than NP exposure, despite the lower total Cu exposure. For genes induced by all Cu exposures, pulse ion treatment led to temporally distinct transcriptional responses (peak inductions generally at 10 or 30 min) compared to both the gradual ion and NP treatments (peak inductions generally at 60 min). This time-resolved depiction of E. coli's transcriptional response to an antimicrobial soluble ENM identifies a response unique to ENM exposure and highlights the importance of sampling time when considering soluble ENM biological impacts. It also underscores the need, in nanotoxicity studies, to consider ionic controls that reflect the slow release of ion from soluble ENMs.
中文翻译:

对CuO纳米颗粒的时间依赖性细菌转录反应与Cu 2+不同,并提供了对CuO纳米颗粒毒性机制的见解†
更好地了解细菌对CuO工程纳米材料(ENM)的时间分辨基因表达反应可能会导致对其抗菌作用的机理有了更好的了解。在这项研究中,逆转录酶定量聚合酶链反应(RT-qPCR)已用于表征暴露于CuO纳米颗粒(NP)(100 mg L -1)后大肠杆菌基因表达的时间序列如铜)。作为与NP暴露的比较,包括了两次溶解的Cu暴露,其浓度与从生长介质中的CuO NPs观察到的溶解相匹配(180分钟后总Cu含量为1 ppm)作为与NP暴露的比较。在脉冲溶解的铜暴露中,在实验开始时添加了1 ppm的Cu。在逐渐溶解的Cu暴露中,将总计1 ppm的溶解Cu分为与观察到的CuO NP溶解相匹配的四个剂量(分别在0、10、30和60分钟)。NP暴露导致诱导膜损伤基因表达,这与高光谱成像(HSI)结果一致,该结果确定了暴露后不久对细胞膜具有较高的NP亲和力。暴露于NP的大肠杆菌在60分钟的时间范围内未诱导出活性氧(ROS)响应基因,其中Cu-(copA,cueO和cusC)和蛋白质损伤基因(cpxP)的表达最多-Cu 2+处理导致的诱导最少。尽管总的铜暴露量较低,但两种铜2+处理(脉冲和逐步添加)均比NP暴露导致更高水平的铜和蛋白质损伤反应性基因诱导。对于由所有铜暴露引起的基因,与渐进式离子和NP处理(通常在60分钟达到峰值)相比,脉冲离子处理导致在时间上截然不同的转录响应(通常在10或30分钟达到峰值)。时间分辨的大肠杆菌描述对抗微生物可溶性ENM的转录反应识别出ENM暴露所特有的反应,并突出了考虑可溶性ENM生物学影响时采样时间的重要性。它还强调了在纳米毒性研究中需要考虑能反映可溶ENM中离子缓慢释放的离子控制。
更新日期:2017-10-20
中文翻译:

对CuO纳米颗粒的时间依赖性细菌转录反应与Cu 2+不同,并提供了对CuO纳米颗粒毒性机制的见解†
更好地了解细菌对CuO工程纳米材料(ENM)的时间分辨基因表达反应可能会导致对其抗菌作用的机理有了更好的了解。在这项研究中,逆转录酶定量聚合酶链反应(RT-qPCR)已用于表征暴露于CuO纳米颗粒(NP)(100 mg L -1)后大肠杆菌基因表达的时间序列如铜)。作为与NP暴露的比较,包括了两次溶解的Cu暴露,其浓度与从生长介质中的CuO NPs观察到的溶解相匹配(180分钟后总Cu含量为1 ppm)作为与NP暴露的比较。在脉冲溶解的铜暴露中,在实验开始时添加了1 ppm的Cu。在逐渐溶解的Cu暴露中,将总计1 ppm的溶解Cu分为与观察到的CuO NP溶解相匹配的四个剂量(分别在0、10、30和60分钟)。NP暴露导致诱导膜损伤基因表达,这与高光谱成像(HSI)结果一致,该结果确定了暴露后不久对细胞膜具有较高的NP亲和力。暴露于NP的大肠杆菌在60分钟的时间范围内未诱导出活性氧(ROS)响应基因,其中Cu-(copA,cueO和cusC)和蛋白质损伤基因(cpxP)的表达最多-Cu 2+处理导致的诱导最少。尽管总的铜暴露量较低,但两种铜2+处理(脉冲和逐步添加)均比NP暴露导致更高水平的铜和蛋白质损伤反应性基因诱导。对于由所有铜暴露引起的基因,与渐进式离子和NP处理(通常在60分钟达到峰值)相比,脉冲离子处理导致在时间上截然不同的转录响应(通常在10或30分钟达到峰值)。时间分辨的大肠杆菌描述对抗微生物可溶性ENM的转录反应识别出ENM暴露所特有的反应,并突出了考虑可溶性ENM生物学影响时采样时间的重要性。它还强调了在纳米毒性研究中需要考虑能反映可溶ENM中离子缓慢释放的离子控制。


























 京公网安备 11010802027423号
京公网安备 11010802027423号