当前位置:
X-MOL 学术
›
Chem. Res. Toxicol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Predicting Organ Toxicity Using in Vitro Bioactivity Data and Chemical Structure
Chemical Research in Toxicology ( IF 4.1 ) Pub Date : 2017-10-09 00:00:00 , DOI: 10.1021/acs.chemrestox.7b00084 Jie Liu 1, 2 , Grace Patlewicz 3 , Antony J. Williams 3 , Russell S. Thomas 3 , Imran Shah 3
Chemical Research in Toxicology ( IF 4.1 ) Pub Date : 2017-10-09 00:00:00 , DOI: 10.1021/acs.chemrestox.7b00084 Jie Liu 1, 2 , Grace Patlewicz 3 , Antony J. Williams 3 , Russell S. Thomas 3 , Imran Shah 3
Affiliation
|
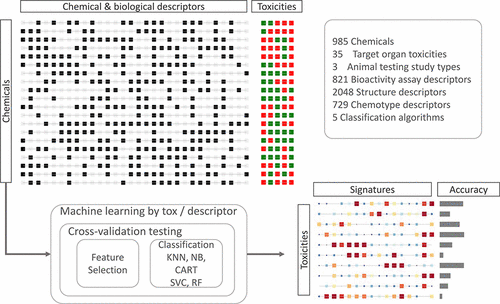
Animal testing alone cannot practically evaluate the health hazard posed by tens of thousands of environmental chemicals. Computational approaches making use of high-throughput experimental data may provide more efficient means to predict chemical toxicity. Here, we use a supervised machine learning strategy to systematically investigate the relative importance of study type, machine learning algorithm, and type of descriptor on predicting in vivo repeat-dose toxicity at the organ-level. A total of 985 compounds were represented using chemical structural descriptors, ToxPrint chemotype descriptors, and bioactivity descriptors from ToxCast in vitro high-throughput screening assays. Using ToxRefDB, a total of 35 target organ outcomes were identified that contained at least 100 chemicals (50 positive and 50 negative). Supervised machine learning was performed using Naïve Bayes, k-nearest neighbor, random forest, classification and regression trees, and support vector classification approaches. Model performance was assessed based on F1 scores using 5-fold cross-validation with balanced bootstrap replicates. Fixed effects modeling showed the variance in F1 scores was explained mostly by target organ outcome, followed by descriptor type, machine learning algorithm, and interactions between these three factors. A combination of bioactivity and chemical structure or chemotype descriptors were the most predictive. Model performance improved with more chemicals (up to a maximum of 24%), and these gains were correlated (ρ = 0.92) with the number of chemicals. Overall, the results demonstrate that a combination of bioactivity and chemical descriptors can accurately predict a range of target organ toxicity outcomes in repeat-dose studies, but specific experimental and methodologic improvements may increase predictivity.
中文翻译:

使用体外生物活性数据和化学结构预测器官毒性
仅动物试验就不能实际评估成千上万种环境化学品对健康的危害。利用高通量实验数据的计算方法可能提供更有效的方法来预测化学毒性。在这里,我们使用有监督的机器学习策略来系统研究研究类型,机器学习算法和描述符类型在预测器官一级体内重复剂量毒性方面的相对重要性。体外使用ToxCast的化学结构描述符,ToxPrint化学型描述符和生物活性描述符代表了985种化合物高通量筛选分析。使用ToxRefDB,总共确定了35种目标器官结局,其中包含至少100种化学物质(50种阳性和50种阴性)。使用朴素贝叶斯,k近邻,随机森林,分类和回归树以及支持向量分类方法进行了监督机器学习。基于F1评分的模型性能通过使用具有平衡自举程序重复的5倍交叉验证进行评估。固定效应模型显示F1分数的差异主要由目标器官的结果解释,其次是描述符类型,机器学习算法以及这三个因素之间的相互作用。生物活性和化学结构或化学型描述符的组合是最可预测的。使用更多的化学药品(最多24%)改善了模型性能,这些收益与化学物质的数量相关(ρ= 0.92)。总体而言,结果表明,在重复剂量研究中,将生物活性和化学描述符结合起来可以准确预测一系列靶器官毒性结果,但是具体的实验和方法改进可能会提高可预测性。
更新日期:2017-10-10
中文翻译:

使用体外生物活性数据和化学结构预测器官毒性
仅动物试验就不能实际评估成千上万种环境化学品对健康的危害。利用高通量实验数据的计算方法可能提供更有效的方法来预测化学毒性。在这里,我们使用有监督的机器学习策略来系统研究研究类型,机器学习算法和描述符类型在预测器官一级体内重复剂量毒性方面的相对重要性。体外使用ToxCast的化学结构描述符,ToxPrint化学型描述符和生物活性描述符代表了985种化合物高通量筛选分析。使用ToxRefDB,总共确定了35种目标器官结局,其中包含至少100种化学物质(50种阳性和50种阴性)。使用朴素贝叶斯,k近邻,随机森林,分类和回归树以及支持向量分类方法进行了监督机器学习。基于F1评分的模型性能通过使用具有平衡自举程序重复的5倍交叉验证进行评估。固定效应模型显示F1分数的差异主要由目标器官的结果解释,其次是描述符类型,机器学习算法以及这三个因素之间的相互作用。生物活性和化学结构或化学型描述符的组合是最可预测的。使用更多的化学药品(最多24%)改善了模型性能,这些收益与化学物质的数量相关(ρ= 0.92)。总体而言,结果表明,在重复剂量研究中,将生物活性和化学描述符结合起来可以准确预测一系列靶器官毒性结果,但是具体的实验和方法改进可能会提高可预测性。



























 京公网安备 11010802027423号
京公网安备 11010802027423号