当前位置:
X-MOL 学术
›
ACS Comb. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Library Design-Facilitated High-Throughput Sequencing of Synthetic Peptide Libraries
ACS Combinatorial Science ( IF 3.903 ) Pub Date : 2017-09-29 00:00:00 , DOI: 10.1021/acscombsci.7b00109 Alexander A. Vinogradov 1 , Zachary P. Gates 1 , Chi Zhang 1 , Anthony J. Quartararo 1 , Kathryn H. Halloran 1 , Bradley L. Pentelute 1
ACS Combinatorial Science ( IF 3.903 ) Pub Date : 2017-09-29 00:00:00 , DOI: 10.1021/acscombsci.7b00109 Alexander A. Vinogradov 1 , Zachary P. Gates 1 , Chi Zhang 1 , Anthony J. Quartararo 1 , Kathryn H. Halloran 1 , Bradley L. Pentelute 1
Affiliation
|
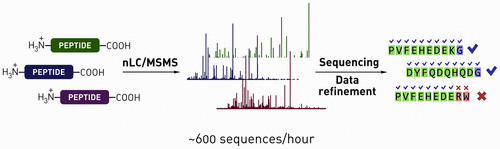
A methodology to achieve high-throughput de novo sequencing of synthetic peptide mixtures is reported. The approach leverages shotgun nanoliquid chromatography coupled with tandem mass spectrometry-based de novo sequencing of library mixtures (up to 2000 peptides) as well as automated data analysis protocols to filter away incorrect assignments, noise, and synthetic side-products. For increasing the confidence in the sequencing results, mass spectrometry-friendly library designs were developed that enabled unambiguous decoding of up to 600 peptide sequences per hour while maintaining greater than 85% sequence identification rates in most cases. The reliability of the reported decoding strategy was additionally confirmed by matching fragmentation spectra for select authentic peptides identified from library sequencing samples. The methods reported here are directly applicable to screening techniques that yield mixtures of active compounds, including particle sorting of one-bead one-compound libraries and affinity enrichment of synthetic library mixtures performed in solution.
中文翻译:

文库设计便利的合成肽库高通量测序
报道了实现合成肽混合物的高通量从头测序的方法。该方法利用shot弹枪纳米液相色谱技术,结合基于串联质谱的从头测序的文库混合物(最多2000个肽段)以及自动数据分析协议,以滤除错误的分配,噪音和合成副产物。为了增加对测序结果的信心,开发了质谱友好的文库设计,该文库设计能够每小时对多达600个肽序列进行明确解码,同时在大多数情况下保持大于85%的序列识别率。通过从文库测序样品中鉴定出的精选真肽的匹配碎片质谱图,还进一步证实了所报道解码策略的可靠性。
更新日期:2017-09-30
中文翻译:

文库设计便利的合成肽库高通量测序
报道了实现合成肽混合物的高通量从头测序的方法。该方法利用shot弹枪纳米液相色谱技术,结合基于串联质谱的从头测序的文库混合物(最多2000个肽段)以及自动数据分析协议,以滤除错误的分配,噪音和合成副产物。为了增加对测序结果的信心,开发了质谱友好的文库设计,该文库设计能够每小时对多达600个肽序列进行明确解码,同时在大多数情况下保持大于85%的序列识别率。通过从文库测序样品中鉴定出的精选真肽的匹配碎片质谱图,还进一步证实了所报道解码策略的可靠性。



























 京公网安备 11010802027423号
京公网安备 11010802027423号