当前位置:
X-MOL 学术
›
Cell Chem. Bio.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader
Cell Chemical Biology ( IF 8.6 ) Pub Date : 2017-11-09 , DOI: 10.1016/j.chembiol.2017.10.005 Hai-Tsang Huang , Dennis Dobrovolsky , Joshiawa Paulk , Guang Yang , Ellen L. Weisberg , Zainab M. Doctor , Dennis L. Buckley , Joong-Heui Cho , Eunhwa Ko , Jaebong Jang , Kun Shi , Hwan Geun Choi , James D. Griffin , Ying Li , Steven P. Treon , Eric S. Fischer , James E. Bradner , Li Tan , Nathanael S. Gray
Cell Chemical Biology ( IF 8.6 ) Pub Date : 2017-11-09 , DOI: 10.1016/j.chembiol.2017.10.005 Hai-Tsang Huang , Dennis Dobrovolsky , Joshiawa Paulk , Guang Yang , Ellen L. Weisberg , Zainab M. Doctor , Dennis L. Buckley , Joong-Heui Cho , Eunhwa Ko , Jaebong Jang , Kun Shi , Hwan Geun Choi , James D. Griffin , Ying Li , Steven P. Treon , Eric S. Fischer , James E. Bradner , Li Tan , Nathanael S. Gray
|
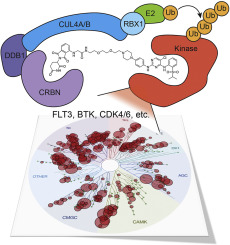
Heterobifunctional molecules that recruit E3 ubiquitin ligases, such as cereblon, for targeted protein degradation represent an emerging pharmacological strategy. A major unanswered question is how generally applicable this strategy is to all protein targets. In this study, we designed a multi-kinase degrader by conjugating a highly promiscuous kinase inhibitor with a cereblon-binding ligand, and used quantitative proteomics to discover 28 kinases, including BTK, PTK2, PTK2B, FLT3, AURKA, AURKB, TEC, ULK1, ITK, and nine members of the CDK family, as degradable. This set of kinases is only a fraction of the intracellular targets bound by the degrader, demonstrating that successful degradation requires more than target engagement. The results guided us to develop selective degraders for FLT3 and BTK, with potentials to improve disease treatment. Together, this study demonstrates an efficient approach to triage a gene family of interest to identify readily degradable targets for further studies and pre-clinical developments.
中文翻译:

一种使用多激酶降解仪查询可降解基因组的化学计量学方法
募集E3泛素连接酶(如cereblon)用于靶向蛋白质降解的异双功能分子代表了一种新兴的药理学策略。一个主要的未解决问题是该策略在所有蛋白质靶标中的适用性。在这项研究中,我们通过将高度混杂的激酶抑制剂与大脑结合配体缀合,设计了多激酶降解剂,并使用定量蛋白质组学发现了28种激酶,包括BTK,PTK2,PTK2B,FLT3,AURKA,AURKB,TEC,ULK1 ,ITK和CDK家族的九名成员,因为它们是可降解的。这组激酶只是降解剂结合的细胞内靶标的一小部分,表明成功的降解需要的不仅仅是靶标参与。该结果指导我们开发了针对FLT3和BTK的选择性降解剂,具有改善疾病治疗的潜力。
更新日期:2018-01-18
中文翻译:

一种使用多激酶降解仪查询可降解基因组的化学计量学方法
募集E3泛素连接酶(如cereblon)用于靶向蛋白质降解的异双功能分子代表了一种新兴的药理学策略。一个主要的未解决问题是该策略在所有蛋白质靶标中的适用性。在这项研究中,我们通过将高度混杂的激酶抑制剂与大脑结合配体缀合,设计了多激酶降解剂,并使用定量蛋白质组学发现了28种激酶,包括BTK,PTK2,PTK2B,FLT3,AURKA,AURKB,TEC,ULK1 ,ITK和CDK家族的九名成员,因为它们是可降解的。这组激酶只是降解剂结合的细胞内靶标的一小部分,表明成功的降解需要的不仅仅是靶标参与。该结果指导我们开发了针对FLT3和BTK的选择性降解剂,具有改善疾病治疗的潜力。



























 京公网安备 11010802027423号
京公网安备 11010802027423号