当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Glycoforest 1.0
Analytical Chemistry ( IF 7.4 ) Pub Date : 2017-09-25 00:00:00 , DOI: 10.1021/acs.analchem.7b02754 Oliver Horlacher 1, 2 , Chunsheng Jin 3 , Davide Alocci 1, 2 , Julien Mariethoz 1, 2 , Markus Müller 1, 2 , Niclas G. Karlsson 3 , Frederique Lisacek 1, 2
Analytical Chemistry ( IF 7.4 ) Pub Date : 2017-09-25 00:00:00 , DOI: 10.1021/acs.analchem.7b02754 Oliver Horlacher 1, 2 , Chunsheng Jin 3 , Davide Alocci 1, 2 , Julien Mariethoz 1, 2 , Markus Müller 1, 2 , Niclas G. Karlsson 3 , Frederique Lisacek 1, 2
Affiliation
|
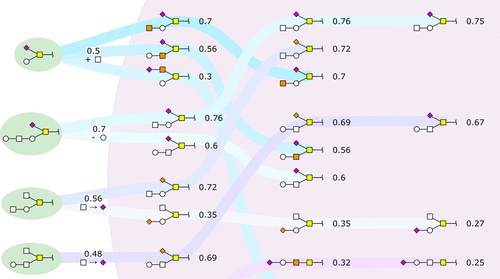
Tandem mass spectrometry, when combined with liquid chromatography and applied to complex mixtures, produces large amounts of raw data, which needs to be analyzed to identify molecular structures. This technique is widely used, particularly in glycomics. Due to a lack of high throughput glycan sequencing software, glycan spectra are predominantly sequenced manually. A challenge for writing glycan-sequencing software is that there is no direct template that can be used to infer structures detectable in an organism. To help alleviate this bottleneck, we present Glycoforest 1.0, a partial de novo algorithm for sequencing glycan structures based on MS/MS spectra. Glycoforest was tested on two data sets (human gastric and salmon mucosa O-linked glycomes) for which MS/MS spectra were annotated manually. Glycoforest generated the human validated structure for 92% of test cases. The correct structure was found as the best scoring match for 70% and among the top 3 matches for 83% of test cases. In addition, the Glycoforest algorithm detected glycan structures from MS/MS spectra missing a manual annotation. In total 1532 MS/MS previously unannotated spectra were annotated by Glycoforest. A portion containing 521 spectra was manually checked confirming that Glycoforest annotated an additional 50 MS/MS spectra overlooked during manual annotation.
中文翻译:

糖森林1.0
串联质谱与液相色谱结合并应用于复杂混合物时,会产生大量原始数据,需要对这些数据进行分析以鉴定分子结构。该技术被广泛使用,尤其是在糖组学中。由于缺乏高通量的聚糖测序软件,因此聚糖谱主要是手动测序。编写聚糖测序软件的挑战在于,没有直接的模板可用于推断生物体中可检测的结构。为了缓解这一瓶颈,我们提出了Glycoforest 1.0,这是一种基于MS / MS光谱对聚糖结构进行测序的部分从头算法。在两个数据集(人胃和鲑鱼粘膜O(MS-MS谱图)手动注释MS / MS谱图。糖森林为92%的测试用例生成了经过人工验证的结构。发现正确的结构是70%的最佳得分匹配项,而83%的测试用例是前三名匹配项。另外,Glycoforest算法从MS / MS光谱中检测到缺少手动注释的聚糖结构。共有1532个MS / MS以前未注释的光谱由Glycoforest注释。手动检查了包含521个光谱的部分,从而确认Glycoforest注释了在手动注释期间被忽略的另外50个MS / MS频谱。
更新日期:2017-09-26
中文翻译:

糖森林1.0
串联质谱与液相色谱结合并应用于复杂混合物时,会产生大量原始数据,需要对这些数据进行分析以鉴定分子结构。该技术被广泛使用,尤其是在糖组学中。由于缺乏高通量的聚糖测序软件,因此聚糖谱主要是手动测序。编写聚糖测序软件的挑战在于,没有直接的模板可用于推断生物体中可检测的结构。为了缓解这一瓶颈,我们提出了Glycoforest 1.0,这是一种基于MS / MS光谱对聚糖结构进行测序的部分从头算法。在两个数据集(人胃和鲑鱼粘膜O(MS-MS谱图)手动注释MS / MS谱图。糖森林为92%的测试用例生成了经过人工验证的结构。发现正确的结构是70%的最佳得分匹配项,而83%的测试用例是前三名匹配项。另外,Glycoforest算法从MS / MS光谱中检测到缺少手动注释的聚糖结构。共有1532个MS / MS以前未注释的光谱由Glycoforest注释。手动检查了包含521个光谱的部分,从而确认Glycoforest注释了在手动注释期间被忽略的另外50个MS / MS频谱。



























 京公网安备 11010802027423号
京公网安备 11010802027423号