当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Scaling behaviour and rate-determining steps in filamentous self-assembly
Chemical Science ( IF 8.4 ) Pub Date : 2017-08-31 00:00:00 , DOI: 10.1039/c7sc01965c Georg Meisl 1, 2, 3, 4 , Luke Rajah 1, 2, 3, 4 , Samuel A. I. Cohen 1, 2, 3, 4 , Manuela Pfammatter 5, 6, 7, 8 , Anđela Šarić 4, 9, 10, 11, 12 , Erik Hellstrand 13, 14, 15, 16 , Alexander K. Buell 17, 18, 19, 20 , Adriano Aguzzi 5, 6, 7, 8 , Sara Linse 13, 14, 15, 16 , Michele Vendruscolo 1, 2, 3, 4 , Christopher M. Dobson 1, 2, 3, 4 , Tuomas P. J. Knowles 1, 2, 3, 4
Chemical Science ( IF 8.4 ) Pub Date : 2017-08-31 00:00:00 , DOI: 10.1039/c7sc01965c Georg Meisl 1, 2, 3, 4 , Luke Rajah 1, 2, 3, 4 , Samuel A. I. Cohen 1, 2, 3, 4 , Manuela Pfammatter 5, 6, 7, 8 , Anđela Šarić 4, 9, 10, 11, 12 , Erik Hellstrand 13, 14, 15, 16 , Alexander K. Buell 17, 18, 19, 20 , Adriano Aguzzi 5, 6, 7, 8 , Sara Linse 13, 14, 15, 16 , Michele Vendruscolo 1, 2, 3, 4 , Christopher M. Dobson 1, 2, 3, 4 , Tuomas P. J. Knowles 1, 2, 3, 4
Affiliation
|
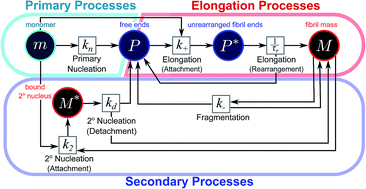
The formation of filaments from naturally occurring protein molecules is a process at the core of a range of functional and aberrant biological phenomena, such as the assembly of the cytoskeleton or the appearance of aggregates in Alzheimer's disease. The macroscopic behaviour associated with such processes is remarkably diverse, ranging from simple nucleated growth to highly cooperative processes with a well-defined lagtime. Thus, conventionally, different molecular mechanisms have been used to explain the self-assembly of different proteins. Here we show that this range of behaviour can be quantitatively captured by a single unifying Petri net that describes filamentous growth in terms of aggregate number and aggregate mass concentrations. By considering general features associated with a particular network connectivity, we are able to establish directly the rate-determining steps of the overall aggregation reaction from the system's scaling behaviour. We illustrate the power of this framework on a range of different experimental and simulated aggregating systems. The approach is general and will be applicable to any future extensions of the reaction network of filamentous self-assembly.
中文翻译:

丝状自组装的定标行为和速率确定步骤
由天然存在的蛋白质分子形成细丝是一系列功能性和异常生物学现象(例如细胞骨架的组装或阿兹海默氏病中聚集体的出现)的核心过程。与此类过程相关的宏观行为非常多样,从简单的有核生长到具有明确定义的滞后时间的高度协作过程。因此,常规上,已经使用不同的分子机制来解释不同蛋白质的自组装。在这里,我们表明,可以通过单个统一的Petri网定量地捕获此行为范围,该网以总数量和总质量浓度描述丝状生长。通过考虑与特定网络连接相关的一般功能,我们能够根据系统的缩放行为直接建立总体聚合反应的速率确定步骤。我们在一系列不同的实验和模拟聚合系统上说明了此框架的强大功能。该方法是通用的,并且将适用于丝状自组装反应网络的任何未来扩展。
更新日期:2017-09-25
中文翻译:

丝状自组装的定标行为和速率确定步骤
由天然存在的蛋白质分子形成细丝是一系列功能性和异常生物学现象(例如细胞骨架的组装或阿兹海默氏病中聚集体的出现)的核心过程。与此类过程相关的宏观行为非常多样,从简单的有核生长到具有明确定义的滞后时间的高度协作过程。因此,常规上,已经使用不同的分子机制来解释不同蛋白质的自组装。在这里,我们表明,可以通过单个统一的Petri网定量地捕获此行为范围,该网以总数量和总质量浓度描述丝状生长。通过考虑与特定网络连接相关的一般功能,我们能够根据系统的缩放行为直接建立总体聚合反应的速率确定步骤。我们在一系列不同的实验和模拟聚合系统上说明了此框架的强大功能。该方法是通用的,并且将适用于丝状自组装反应网络的任何未来扩展。



























 京公网安备 11010802027423号
京公网安备 11010802027423号