当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Optimal preprocessing of serum and urine metabolomic data fusion for staging prostate cancer through design of experiment
Analytica Chimica Acta ( IF 6.2 ) Pub Date : 2017-10-01 , DOI: 10.1016/j.aca.2017.09.019 Hong Zheng , Aimin Cai , Qi Zhou , Pengtao Xu , Liangcai Zhao , Chen Li , Baijun Dong , Hongchang Gao
Analytica Chimica Acta ( IF 6.2 ) Pub Date : 2017-10-01 , DOI: 10.1016/j.aca.2017.09.019 Hong Zheng , Aimin Cai , Qi Zhou , Pengtao Xu , Liangcai Zhao , Chen Li , Baijun Dong , Hongchang Gao
|
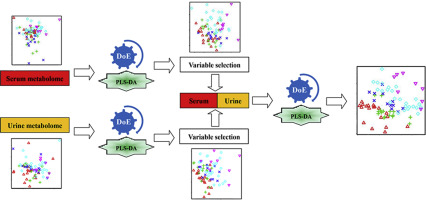
Accurate classification of cancer stages will achieve precision treatment for cancer. Metabolomics presents biological phenotypes at the metabolite level and holds a great potential for cancer classification. Since metabolomic data can be obtained from different samples or analytical techniques, data fusion has been applied to improve classification accuracy. Data preprocessing is an essential step during metabolomic data analysis. Therefore, we developed an innovative optimization method to select a proper data preprocessing strategy for metabolomic data fusion using a design of experiment approach for improving the classification of prostate cancer (PCa) stages. In this study, urine and serum samples were collected from participants at five phases of PCa and analyzed using a 1H NMR-based metabolomic approach. Partial least squares-discriminant analysis (PLS-DA) was used as a classification model and its performance was assessed by goodness of fit (R2) and predictive ability (Q2). Results show that data preprocessing significantly affect classification performance and depends on data properties. Using the fused metabolomic data from urine and serum, PLS-DA model with the optimal data preprocessing (R2 = 0.729, Q2 = 0.504, P < 0.0001) can effectively improve model performance and achieve a better classification result for PCa stages as compared with that without data preprocessing (R2 = 0.139, Q2 = 0.006, P = 0.450). Therefore, we propose that metabolomic data fusion integrated with an optimal data preprocessing strategy can significantly improve the classification of cancer stages for precision treatment.
中文翻译:

通过实验设计对血清和尿液代谢组学数据融合进行优化预处理用于前列腺癌分期
准确分类癌症分期,实现癌症精准治疗。代谢组学在代谢物水平上呈现生物表型,在癌症分类方面具有巨大潜力。由于代谢组学数据可以从不同的样本或分析技术中获得,因此数据融合已被应用于提高分类精度。数据预处理是代谢组学数据分析过程中必不可少的步骤。因此,我们开发了一种创新的优化方法,使用改进前列腺癌 (PCa) 分期分类的实验方法设计,为代谢组学数据融合选择合适的数据预处理策略。在这项研究中,从参与者的五个 PCa 阶段收集尿液和血清样本,并使用基于 1H NMR 的代谢组学方法进行分析。偏最小二乘判别分析 (PLS-DA) 用作分类模型,其性能通过拟合优度 (R2) 和预测能力 (Q2) 进行评估。结果表明,数据预处理显着影响分类性能并取决于数据属性。使用尿液和血清的融合代谢组学数据,PLS-DA模型与最佳数据预处理(R2 = 0.729,Q2 = 0.504,P < 0.0001)相比,可以有效提高模型性能并获得更好的PCa分期分类结果没有数据预处理(R2 = 0.139,Q2 = 0.006,P = 0.450)。因此,我们建议将代谢组学数据融合与最佳数据预处理策略相结合,可以显着改善癌症分期的分类,以实现精准治疗。
更新日期:2017-10-01
中文翻译:

通过实验设计对血清和尿液代谢组学数据融合进行优化预处理用于前列腺癌分期
准确分类癌症分期,实现癌症精准治疗。代谢组学在代谢物水平上呈现生物表型,在癌症分类方面具有巨大潜力。由于代谢组学数据可以从不同的样本或分析技术中获得,因此数据融合已被应用于提高分类精度。数据预处理是代谢组学数据分析过程中必不可少的步骤。因此,我们开发了一种创新的优化方法,使用改进前列腺癌 (PCa) 分期分类的实验方法设计,为代谢组学数据融合选择合适的数据预处理策略。在这项研究中,从参与者的五个 PCa 阶段收集尿液和血清样本,并使用基于 1H NMR 的代谢组学方法进行分析。偏最小二乘判别分析 (PLS-DA) 用作分类模型,其性能通过拟合优度 (R2) 和预测能力 (Q2) 进行评估。结果表明,数据预处理显着影响分类性能并取决于数据属性。使用尿液和血清的融合代谢组学数据,PLS-DA模型与最佳数据预处理(R2 = 0.729,Q2 = 0.504,P < 0.0001)相比,可以有效提高模型性能并获得更好的PCa分期分类结果没有数据预处理(R2 = 0.139,Q2 = 0.006,P = 0.450)。因此,我们建议将代谢组学数据融合与最佳数据预处理策略相结合,可以显着改善癌症分期的分类,以实现精准治疗。


























 京公网安备 11010802027423号
京公网安备 11010802027423号