当前位置:
X-MOL 学术
›
Bioconjugate Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Selective Binders of the Tandem Src Homology 2 Domains in Syk and Zap70 Protein Kinases by DNA-Programmed Spatial Screening
Bioconjugate Chemistry ( IF 4.7 ) Pub Date : 2017-08-30 00:00:00 , DOI: 10.1021/acs.bioconjchem.7b00382 Michaela Marczynke 1 , Katharina Gröger 1 , Oliver Seitz 1
Bioconjugate Chemistry ( IF 4.7 ) Pub Date : 2017-08-30 00:00:00 , DOI: 10.1021/acs.bioconjchem.7b00382 Michaela Marczynke 1 , Katharina Gröger 1 , Oliver Seitz 1
Affiliation
|
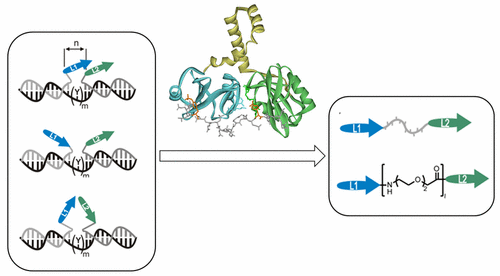
Members of the Syk family of tyrosine kinases arrange Src homology 2 (SH2) domains in tandem to allow the firm binding of immunoreceptor tyrosine-based interaction motifs (ITAMs). While the advantages provided by the bivalency enhanced interactions are evident, the impact on binding specificity is less-clear. For example, the spleen tyrosine kinase (Syk) and the ζ-chain-associated protein kinase (ZAP-70) recognize the consensus sequence pYXXI/L(X)6–8 pYXXI/L with near-identical nanomolar affinity. The nondiscriminatory recognition, on the one hand, poses a specificity challenge for the design of subtype selective protein binders and, on the other hand, raises the question as to how differential activation of Syk and ZAP-70 is ensured when both kinases are co-expressed. Herein, we identified the criteria for the design of binders that specifically address either the Syk or the Zap-70 tSH2 domain. Our approach is based on DNA-programmed spatial screening. Tyrosine-phosphorylated peptides containing the pYXXI/L motif were attached to oligonucleotides and aligned in tandem on a DNA template by means of nucleic acid hybridization. The distance between the pYXXI/L motifs and the orientation of strands were varied. The exploration exposed remarkably different recognition characteristics. While Syk tSH2 has a rather broad substrate scope, ZAP-70 tSH2 required a proximal arrangement of the phosphotyrosine ligands in defined strand orientation. The spatial screen led to the design of mutually selective, DNA-free binders, which discriminate Zap-70 and Syk tSH2 by 1 order of magnitude in affinity.
中文翻译:

Syk和Zap70蛋白激酶的串联Src同源2域的选择性结合剂的DNA程序设计的空间筛选。
Syk酪氨酸激酶家族的成员串联排列Src同源性2(SH2)域,以使免疫受体基于酪氨酸的相互作用基序(ITAM)牢固结合。虽然双价增强相互作用提供的优势是显而易见的,但对结合特异性的影响尚不清楚。例如,脾酪氨酸激酶(Syk)和ζ链相关蛋白激酶(ZAP-70)识别共有序列pYXXI / L(X)6-8pYXXI / L具有几乎相同的纳摩尔亲和力。一方面,非歧视性识别对亚型选择性蛋白结合剂的设计提出了特异性挑战,另一方面,提出了一个问题,即当两种激酶共同参与时,如何确保Syk和ZAP-70的差异激活。表达。在这里,我们确定了专门针对Syk或Zap-70 tSH2域的粘合剂设计标准。我们的方法基于DNA编程的空间筛选。将含有pYXXI / L基序的酪氨酸磷酸化肽连接至寡核苷酸,并通过核酸杂交将其串联排列在DNA模板上。pYXXI / L基序之间的距离和链的方向是变化的。探索揭示了明显不同的识别特征。尽管Syk tSH2具有相当宽的底物范围,但ZAP-70 tSH2要求磷酸酪氨酸配体以确定的链取向近端排列。空间筛选导致了相互选择性,不含DNA的结合剂的设计,该结合剂将Zap-70和Syk tSH2的亲和力区分了1个数量级。
更新日期:2017-08-30
中文翻译:

Syk和Zap70蛋白激酶的串联Src同源2域的选择性结合剂的DNA程序设计的空间筛选。
Syk酪氨酸激酶家族的成员串联排列Src同源性2(SH2)域,以使免疫受体基于酪氨酸的相互作用基序(ITAM)牢固结合。虽然双价增强相互作用提供的优势是显而易见的,但对结合特异性的影响尚不清楚。例如,脾酪氨酸激酶(Syk)和ζ链相关蛋白激酶(ZAP-70)识别共有序列pYXXI / L(X)6-8pYXXI / L具有几乎相同的纳摩尔亲和力。一方面,非歧视性识别对亚型选择性蛋白结合剂的设计提出了特异性挑战,另一方面,提出了一个问题,即当两种激酶共同参与时,如何确保Syk和ZAP-70的差异激活。表达。在这里,我们确定了专门针对Syk或Zap-70 tSH2域的粘合剂设计标准。我们的方法基于DNA编程的空间筛选。将含有pYXXI / L基序的酪氨酸磷酸化肽连接至寡核苷酸,并通过核酸杂交将其串联排列在DNA模板上。pYXXI / L基序之间的距离和链的方向是变化的。探索揭示了明显不同的识别特征。尽管Syk tSH2具有相当宽的底物范围,但ZAP-70 tSH2要求磷酸酪氨酸配体以确定的链取向近端排列。空间筛选导致了相互选择性,不含DNA的结合剂的设计,该结合剂将Zap-70和Syk tSH2的亲和力区分了1个数量级。



























 京公网安备 11010802027423号
京公网安备 11010802027423号