当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Linking High-Throughput Screens to Identify MoAs and Novel Inhibitors of Mycobacterium tuberculosis Dihydrofolate Reductase
ACS Chemical Biology ( IF 4 ) Pub Date : 2017-08-29 00:00:00 , DOI: 10.1021/acschembio.7b00468 John P. Santa Maria 1 , Yumi Park 2 , Lihu Yang 3 , Nicholas Murgolo 4 , Michael D. Altman 1 , Paul Zuck 5 , Greg Adam 6 , Chad Chamberlin 7 , Peter Saradjian 7 , Peter Dandliker 7 , Helena I. M. Boshoff 2 , Clifton E. Barry 2 , Charles Garlisi 8 , David B. Olsen 9 , Katherine Young 9 , Meir Glick 1 , Elliott Nickbarg 7 , Peter S. Kutchukian 1
ACS Chemical Biology ( IF 4 ) Pub Date : 2017-08-29 00:00:00 , DOI: 10.1021/acschembio.7b00468 John P. Santa Maria 1 , Yumi Park 2 , Lihu Yang 3 , Nicholas Murgolo 4 , Michael D. Altman 1 , Paul Zuck 5 , Greg Adam 6 , Chad Chamberlin 7 , Peter Saradjian 7 , Peter Dandliker 7 , Helena I. M. Boshoff 2 , Clifton E. Barry 2 , Charles Garlisi 8 , David B. Olsen 9 , Katherine Young 9 , Meir Glick 1 , Elliott Nickbarg 7 , Peter S. Kutchukian 1
Affiliation
|
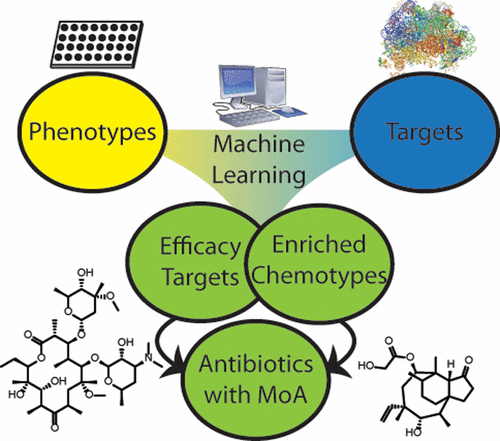
Though phenotypic and target-based high-throughput screening approaches have been employed to discover new antibiotics, the identification of promising therapeutic candidates remains challenging. Each approach provides different information, and understanding their results can provide hypotheses for a mechanism of action (MoA) and reveal actionable chemical matter. Here, we describe a framework for identifying efficacy targets of bioactive compounds. High throughput biophysical profiling against a broad range of targets coupled with machine learning was employed to identify chemical features with predicted efficacy targets for a given phenotypic screen. We validate the approach on data from a set of 55 000 compounds in 24 historical internal antibacterial phenotypic screens and 636 bacterial targets screened in high-throughput biophysical binding assays. Models were built to reveal the relationships between phenotype, target, and chemotype, which recapitulated mechanisms for known antibacterials. We also prospectively identified novel inhibitors of dihydrofolate reductase with nanomolar antibacterial efficacy against Mycobacterium tuberculosis. Molecular modeling provided structural insight into target–ligand interactions underlying selective killing activity toward mycobacteria over human cells.
中文翻译:

链接高通量屏幕以鉴定结核分枝杆菌二氢叶酸还原酶的MoAs和新型抑制剂
尽管已经采用了基于表型和靶标的高通量筛选方法来发现新的抗生素,但是鉴定有前途的候选治疗药物仍然具有挑战性。每种方法都提供不同的信息,了解它们的结果可以为作用机理(MoA)提供假设,并揭示可操作的化学物质。在这里,我们描述了用于识别生物活性化合物功效目标的框架。针对各种靶标的高通量生物物理特征分析以及机器学习被用于识别具有给定表型筛选的预期功效靶标的化学特征。我们在24种历史内部抗菌表型筛选和636种细菌靶标(通过高通量生物物理结合测定法筛选)中,对来自55 000个化合物的数据集上的方法进行了验证。建立模型以揭示表型,靶标和化学型之间的关系,这些关系概括了已知抗菌剂的机制。我们还前瞻性地确定了新型的二氢叶酸还原酶抑制剂,具有纳摩尔抗细菌功效。结核分枝杆菌。分子建模提供了对靶标-配体相互作用的结构性见解,这些靶标-配体之间的相互作用是对人类细胞对分枝杆菌的选择性杀伤活性的基础。
更新日期:2017-08-29
中文翻译:

链接高通量屏幕以鉴定结核分枝杆菌二氢叶酸还原酶的MoAs和新型抑制剂
尽管已经采用了基于表型和靶标的高通量筛选方法来发现新的抗生素,但是鉴定有前途的候选治疗药物仍然具有挑战性。每种方法都提供不同的信息,了解它们的结果可以为作用机理(MoA)提供假设,并揭示可操作的化学物质。在这里,我们描述了用于识别生物活性化合物功效目标的框架。针对各种靶标的高通量生物物理特征分析以及机器学习被用于识别具有给定表型筛选的预期功效靶标的化学特征。我们在24种历史内部抗菌表型筛选和636种细菌靶标(通过高通量生物物理结合测定法筛选)中,对来自55 000个化合物的数据集上的方法进行了验证。建立模型以揭示表型,靶标和化学型之间的关系,这些关系概括了已知抗菌剂的机制。我们还前瞻性地确定了新型的二氢叶酸还原酶抑制剂,具有纳摩尔抗细菌功效。结核分枝杆菌。分子建模提供了对靶标-配体相互作用的结构性见解,这些靶标-配体之间的相互作用是对人类细胞对分枝杆菌的选择性杀伤活性的基础。



























 京公网安备 11010802027423号
京公网安备 11010802027423号