Molecular Cell ( IF 16.0 ) Pub Date : 2017-07-20 , DOI: 10.1016/j.molcel.2017.06.027 Arnaud R. Krebs , Dilek Imanci , Leslie Hoerner , Dimos Gaidatzis , Lukas Burger , Dirk Schübeler
|
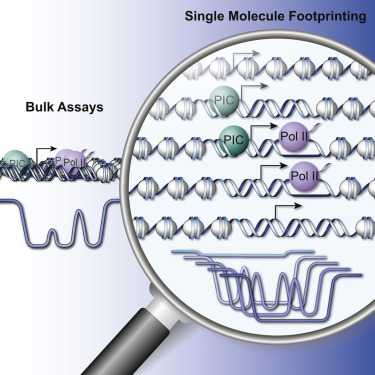
Transcription initiation entails chromatin opening followed by pre-initiation complex formation and RNA polymerase II recruitment. Subsequent polymerase elongation requires additional signals, resulting in increased residence time downstream of the start site, a phenomenon referred to as pausing. Here, we harnessed single-molecule footprinting to quantify distinct steps of initiation in vivo throughout the Drosophila genome. This identifies the impact of promoter structure on initiation dynamics in relation to nucleosomal occupancy. Additionally, perturbation of transcriptional initiation reveals an unexpectedly high turnover of polymerases at paused promoters—an observation confirmed at the level of nascent RNAs. These observations argue that absence of elongation is largely caused by premature termination rather than by stable polymerase stalling. In support of this non-processive model, we observe that induction of the paused heat shock promoter depends on continuous initiation. Our study provides a framework to quantify protein binding at single-molecule resolution and refines concepts of transcriptional pausing.
中文翻译:

全基因组的单分子足迹揭示了在暂停的启动子高RNA聚合酶II营业额。
转录起始需要染色质开放,然后起始前复合物的形成和RNA聚合酶II的募集。随后的聚合酶延伸需要附加信号,导致起始位点下游的停留时间增加,这种现象称为暂停。在这里,我们利用单分子足迹来定量整个果蝇体内的不同起始步骤。基因组。这确定了启动子结构对与核小体占用有关的启动动力学的影响。此外,转录起始的扰动揭示了在暂停的启动子处聚合酶的出乎意料的高周转率-在新生RNA的水平上证实了这一观察结果。这些观察结果认为,伸长率的缺乏主要是由于过早终止而不是由稳定的聚合酶停滞引起的。为了支持该非过程模型,我们观察到了暂停的热激启动子的诱导取决于连续的引发。我们的研究提供了一个框架,可以以单分子分辨率量化蛋白质结合并完善转录暂停的概念。



























 京公网安备 11010802027423号
京公网安备 11010802027423号