当前位置:
X-MOL 学术
›
Mol. Biosyst.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
pLoc-mPlant: predict subcellular localization of multi-location plant proteins by incorporating the optimal GO information into general PseAAC
Molecular BioSystems Pub Date : 2017-07-12 00:00:00 , DOI: 10.1039/c7mb00267j Xiang Cheng 1, 2, 3, 4 , Xuan Xiao 1, 2, 3, 4, 5 , Kuo-Chen Chou 5, 6, 7, 8, 9
Molecular BioSystems Pub Date : 2017-07-12 00:00:00 , DOI: 10.1039/c7mb00267j Xiang Cheng 1, 2, 3, 4 , Xuan Xiao 1, 2, 3, 4, 5 , Kuo-Chen Chou 5, 6, 7, 8, 9
Affiliation
|
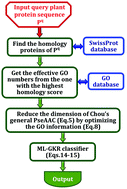
One of the fundamental goals in cellular biochemistry is to identify the functions of proteins in the context of compartments that organize them in the cellular environment. To realize this, it is indispensable to develop an automated method for fast and accurate identification of the subcellular locations of uncharacterized proteins. The current study is focused on plant protein subcellular location prediction based on the sequence information alone. Although considerable efforts have been made in this regard, the problem is far from being solved yet. Most of the existing methods can be used to deal with single-location proteins only. Actually, proteins with multi-locations may have some special biological functions. This kind of multiplex protein is particularly important for both basic research and drug design. Using the multi-label theory, we present a new predictor called “pLoc-mPlant” by extracting the optimal GO (Gene Ontology) information into the Chou's general PseAAC (Pseudo Amino Acid Composition). Rigorous cross-validation on the same stringent benchmark dataset indicated that the proposed pLoc-mPlant predictor is remarkably superior to iLoc-Plant, the state-of-the-art method for predicting plant protein subcellular localization. To maximize the convenience of most experimental scientists, a user-friendly web-server for the new predictor has been established at http://www.jci-bioinfo.cn/pLoc-mPlant/, by which users can easily get their desired results without the need to go through the complicated mathematics involved.
中文翻译:

pLoc-mPlant:通过将最佳GO信息整合到一般的PseAAC中,预测多位置植物蛋白的亚细胞定位
细胞生物化学的基本目标之一是确定在细胞环境中组织蛋白质的区域中蛋白质的功能。为了实现这一点,必须开发一种自动方法来快速准确地鉴定未表征蛋白的亚细胞位置。当前的研究集中在仅基于序列信息的植物蛋白亚细胞定位预测上。尽管在这方面已经做出了很大的努力,但是该问题仍未解决。现有的大多数方法只能用于处理单位置蛋白。实际上,具有多个位置的蛋白质可能具有某些特殊的生物学功能。这种多重蛋白对于基础研究和药物设计都特别重要。利用多标签理论,通过将最佳的GO(基因本体论)信息提取到Chou的一般PseAAC(伪氨基酸组成)中,我们提出了一种称为“ pLoc-mPlant”的新预测因子。在相同的严格基准数据集上进行严格的交叉验证表明,提出的pLoc-mPlant预测因子明显优于iLoc-Plant,后者是预测植物蛋白亚细胞定位的最新技术。为了最大程度地提高大多数实验科学家的便利性,已在http://www.jci-bioinfo.cn/pLoc-mPlant/上建立了用于新预测变量的用户友好型Web服务器,用户可以通过该服务器轻松获得所需的结果。无需进行复杂的数学运算。一般的PseAAC(伪氨基酸组成)。在相同的严格基准数据集上进行严格的交叉验证表明,提出的pLoc-mPlant预测因子明显优于iLoc-Plant,后者是预测植物蛋白亚细胞定位的最新技术。为了最大程度地提高大多数实验科学家的便利性,已在http://www.jci-bioinfo.cn/pLoc-mPlant/上建立了用于新预测变量的用户友好型Web服务器,用户可以通过该服务器轻松获得所需的结果。无需进行复杂的数学运算。一般的PseAAC(伪氨基酸组成)。在相同的严格基准数据集上进行严格的交叉验证表明,提出的pLoc-mPlant预测因子明显优于iLoc-Plant,后者是预测植物蛋白亚细胞定位的最新技术。为了最大程度地提高大多数实验科学家的便利性,已在http://www.jci-bioinfo.cn/pLoc-mPlant/上建立了用于新预测变量的用户友好型Web服务器,用户可以通过该服务器轻松获得所需的结果。无需进行复杂的数学运算。
更新日期:2017-08-22
中文翻译:

pLoc-mPlant:通过将最佳GO信息整合到一般的PseAAC中,预测多位置植物蛋白的亚细胞定位
细胞生物化学的基本目标之一是确定在细胞环境中组织蛋白质的区域中蛋白质的功能。为了实现这一点,必须开发一种自动方法来快速准确地鉴定未表征蛋白的亚细胞位置。当前的研究集中在仅基于序列信息的植物蛋白亚细胞定位预测上。尽管在这方面已经做出了很大的努力,但是该问题仍未解决。现有的大多数方法只能用于处理单位置蛋白。实际上,具有多个位置的蛋白质可能具有某些特殊的生物学功能。这种多重蛋白对于基础研究和药物设计都特别重要。利用多标签理论,通过将最佳的GO(基因本体论)信息提取到Chou的一般PseAAC(伪氨基酸组成)中,我们提出了一种称为“ pLoc-mPlant”的新预测因子。在相同的严格基准数据集上进行严格的交叉验证表明,提出的pLoc-mPlant预测因子明显优于iLoc-Plant,后者是预测植物蛋白亚细胞定位的最新技术。为了最大程度地提高大多数实验科学家的便利性,已在http://www.jci-bioinfo.cn/pLoc-mPlant/上建立了用于新预测变量的用户友好型Web服务器,用户可以通过该服务器轻松获得所需的结果。无需进行复杂的数学运算。一般的PseAAC(伪氨基酸组成)。在相同的严格基准数据集上进行严格的交叉验证表明,提出的pLoc-mPlant预测因子明显优于iLoc-Plant,后者是预测植物蛋白亚细胞定位的最新技术。为了最大程度地提高大多数实验科学家的便利性,已在http://www.jci-bioinfo.cn/pLoc-mPlant/上建立了用于新预测变量的用户友好型Web服务器,用户可以通过该服务器轻松获得所需的结果。无需进行复杂的数学运算。一般的PseAAC(伪氨基酸组成)。在相同的严格基准数据集上进行严格的交叉验证表明,提出的pLoc-mPlant预测因子明显优于iLoc-Plant,后者是预测植物蛋白亚细胞定位的最新技术。为了最大程度地提高大多数实验科学家的便利性,已在http://www.jci-bioinfo.cn/pLoc-mPlant/上建立了用于新预测变量的用户友好型Web服务器,用户可以通过该服务器轻松获得所需的结果。无需进行复杂的数学运算。



























 京公网安备 11010802027423号
京公网安备 11010802027423号